+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asp-bound GltPh RSMR mutant in IFS-A2 state | |||||||||
 Map data Map data | Asp-bound GltPh RSMR mutant in IFS state | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationL-aspartate transmembrane transport / amino acid:sodium symporter activity / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / chloride transmembrane transporter activity / protein homotrimerization / chloride transmembrane transport / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Pyrococcus horikoshii (archaea) Pyrococcus horikoshii (archaea) | |||||||||
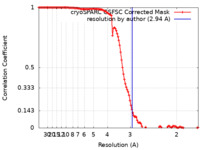
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Huang Y / Boudker O | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2023 Journal: J Am Chem Soc / Year: 2023Title: Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by F NMR and Cryo-EM. Authors: Yun Huang / Krishna D Reddy / Clay Bracken / Biao Qiu / Wenhu Zhan / David Eliezer / Olga Boudker /  Abstract: Limited chemical shift dispersion represents a significant barrier to studying multistate equilibria of large membrane proteins by F NMR. We describe a novel monofluoroethyl F probe that dramatically ...Limited chemical shift dispersion represents a significant barrier to studying multistate equilibria of large membrane proteins by F NMR. We describe a novel monofluoroethyl F probe that dramatically increases the chemical shift dispersion. The improved conformational sensitivity and line shape enable the detection of previously unresolved states in one-dimensional (1D) F NMR spectra of a 134 kDa membrane transporter. Changes in the populations of these states in response to ligand binding, mutations, and temperature correlate with population changes of distinct conformations in structural ensembles determined by single-particle cryo-electron microscopy (cryo-EM). Thus, F NMR can guide sample preparation to discover and visualize novel conformational states and facilitate image analysis and three-dimensional (3D) classification. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26497.map.gz emd_26497.map.gz | 83.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26497-v30.xml emd-26497-v30.xml emd-26497.xml emd-26497.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26497_fsc.xml emd_26497_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26497.png emd_26497.png | 107.2 KB | ||
| Others |  emd_26497_half_map_1.map.gz emd_26497_half_map_1.map.gz emd_26497_half_map_2.map.gz emd_26497_half_map_2.map.gz | 154.6 MB 154.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26497 http://ftp.pdbj.org/pub/emdb/structures/EMD-26497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26497 | HTTPS FTP |
-Related structure data
| Related structure data |  7ugvMC  7ugxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26497.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26497.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asp-bound GltPh RSMR mutant in IFS state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.852 Å | ||||||||||||||||||||||||||||||||||||
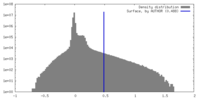
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_26497_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
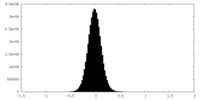
| Density Histograms |
-Half map: Half Map 2
| File | emd_26497_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
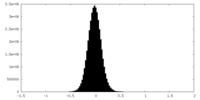
| Density Histograms |
- Sample components
Sample components
-Entire : GltPh
| Entire | Name: GltPh |
|---|---|
| Components |
|
-Supramolecule #1: GltPh
| Supramolecule | Name: GltPh / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Pyrococcus horikoshii (archaea) Pyrococcus horikoshii (archaea) |
-Macromolecule #1: Glutamate transporter homolog
| Macromolecule | Name: Glutamate transporter homolog / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus horikoshii (archaea) Pyrococcus horikoshii (archaea)Strain: ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3 |
| Molecular weight | Theoretical: 44.13323 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGLYRKYIEY PVLQKILIGL ILGAIVGLIL GHYGYAHAVH TYVKPFGDLF VRLLKMLVMP IVFASLVVGA ASISPARLGR VGVKIVVYY LLTSAFAVTL GIIMARLFNP GAGIHLAVGG QQFQPHQAPP LVHILLDIVP TNPFGALANG QVLPTIFFAI I LGIAITYL ...String: MGLYRKYIEY PVLQKILIGL ILGAIVGLIL GHYGYAHAVH TYVKPFGDLF VRLLKMLVMP IVFASLVVGA ASISPARLGR VGVKIVVYY LLTSAFAVTL GIIMARLFNP GAGIHLAVGG QQFQPHQAPP LVHILLDIVP TNPFGALANG QVLPTIFFAI I LGIAITYL MNSENEKVRK SAETLLDAIN GLAEAMYKIV NGVMQYAPIG VFALIAHVMA HQGVHVVGEL AKVTAAVYVG LT LQILLVY FVLLKIYGID PISFIKHAKD AMLTAFVTSS SSGTLPVTMR VAKEMGISEG IYSFTLPLGA TINMDGTALY QGV ATFFIA NALGSHLTVG QQLTIVLTAV LASIGTAGVP GAGAIMLAMV LHSVGLPLTD PNVAAAYA(EFC)I LGIDAILDRG RTMVNVTGD LTGTAIVAKT EGT |
-Macromolecule #2: ASPARTIC ACID
| Macromolecule | Name: ASPARTIC ACID / type: ligand / ID: 2 / Number of copies: 1 / Formula: ASP |
|---|---|
| Molecular weight | Theoretical: 133.103 Da |
| Chemical component information |  ChemComp-ASP: |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 3 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 303 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.94 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)