[English] 日本語
 Yorodumi
Yorodumi- EMDB-26488: Cryo-EM structure of TRPV3 in complex with the anesthetic dyclonine -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TRPV3 in complex with the anesthetic dyclonine | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Transient Receptor Potential V Family Member 3 / TRP / channel / closed / TRPV3 / anesthetic / dyclonine / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of hair cycle / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / sodium channel activity / monoatomic ion channel activity / monoatomic cation channel activity / calcium channel activity / lysosome / receptor complex ...negative regulation of hair cycle / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / sodium channel activity / monoatomic ion channel activity / monoatomic cation channel activity / calcium channel activity / lysosome / receptor complex / metal ion binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | ||||||||||||||||||
 Authors Authors | Neuberger A / Nadezhdin KD / Sobolevsky AI | ||||||||||||||||||
| Funding support |  United States, United States,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural mechanism of TRPV3 channel inhibition by the anesthetic dyclonine. Authors: Arthur Neuberger / Kirill D Nadezhdin / Alexander I Sobolevsky /  Abstract: Skin diseases are common human illnesses that occur in all cultures, at all ages, and affect between 30% and 70% of individuals globally. TRPV3 is a cation-permeable TRP channel predominantly ...Skin diseases are common human illnesses that occur in all cultures, at all ages, and affect between 30% and 70% of individuals globally. TRPV3 is a cation-permeable TRP channel predominantly expressed in skin keratinocytes, implicated in cutaneous sensation and associated with numerous skin diseases. TRPV3 is inhibited by the local anesthetic dyclonine, traditionally used for topical applications to relieve pain and itch. However, the structural basis of TRPV3 inhibition by dyclonine has remained elusive. Here we present a cryo-EM structure of a TRPV3-dyclonine complex that reveals binding of the inhibitor in the portals which connect the membrane environment surrounding the channel to the central cavity of the channel pore. We propose a mechanism of TRPV3 inhibition in which dyclonine molecules stick out into the channel pore, creating a barrier for ion conductance. The allosteric binding site of dyclonine can serve as a template for the design of new TRPV3-targeting drugs. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26488.map.gz emd_26488.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26488-v30.xml emd-26488-v30.xml emd-26488.xml emd-26488.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26488.png emd_26488.png | 86.7 KB | ||
| Filedesc metadata |  emd-26488.cif.gz emd-26488.cif.gz | 6.7 KB | ||
| Others |  emd_26488_half_map_1.map.gz emd_26488_half_map_1.map.gz emd_26488_half_map_2.map.gz emd_26488_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26488 http://ftp.pdbj.org/pub/emdb/structures/EMD-26488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26488 | HTTPS FTP |
-Validation report
| Summary document |  emd_26488_validation.pdf.gz emd_26488_validation.pdf.gz | 893.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26488_full_validation.pdf.gz emd_26488_full_validation.pdf.gz | 893 KB | Display | |
| Data in XML |  emd_26488_validation.xml.gz emd_26488_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_26488_validation.cif.gz emd_26488_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26488 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26488 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26488 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26488 | HTTPS FTP |
-Related structure data
| Related structure data |  7uggMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26488.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26488.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_26488_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
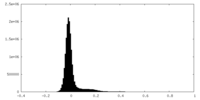
| Density Histograms |
-Half map: #2
| File | emd_26488_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
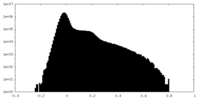
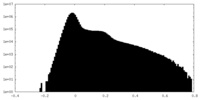
| Density Histograms |
- Sample components
Sample components
-Entire : sample 1
| Entire | Name: sample 1 |
|---|---|
| Components |
|
-Supramolecule #1: sample 1
| Supramolecule | Name: sample 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.47 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 3
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 3 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.573641 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAHSKEMAP LMGKRTTAPG GNPVVLTEKR PADLTPTKKS AHFFLEIEGF EPNPTVTKTS PPIFSKPMDS NIRQCLSGNC DDMDSPQSP QDDVTETPSN PNSPSANLAK EEQRQKKKRL KKRIFAAVSE GCVEELRELL QDLQDLCRRR RGLDVPDFLM H KLTASDTG ...String: MGAHSKEMAP LMGKRTTAPG GNPVVLTEKR PADLTPTKKS AHFFLEIEGF EPNPTVTKTS PPIFSKPMDS NIRQCLSGNC DDMDSPQSP QDDVTETPSN PNSPSANLAK EEQRQKKKRL KKRIFAAVSE GCVEELRELL QDLQDLCRRR RGLDVPDFLM H KLTASDTG KTCLMKALLN INPNTKEIVR ILLAFAEEND ILDRFINAEY TEEAYEGQTA LNIAIERRQG DITAVLIAAG AD VNAHAKG VFFNPKYQHE GFYFGETPLA LAACTNQPEI VQLLMENEQT DITSQDSRGN NILHALVTVA EDFKTQNDFV KRM YDMILL RSGNWELETM RNNDGLTPLQ LAAKMGKAEI LKYILSREIK EKPLRSLSRK FTDWAYGPVS SSLYDLTNVD TTTD NSVLE IIVYNTNIDN RHEMLTLEPL HTLLHTKWKK FAKYMFFLSF CFYFFYNITL TLVSYYRPRE DEDLPHPLAL THKMS WLQL LGRMFVLIWA TCISVKEGIA IFLLRPSDLQ SILSDAWFHF VFFVQAVLVI LSVFLYLFAY KEYLACLVLA MALGWA NML YYTRGFQSMG MYSVMIQKVI LHDVLKFLFV YILFLLGFGV ALASLIEKCS KDKKDCSSYG SFSDAVLELF KLTIGLG DL NIQQNSTYPI LFLFLLITYV ILTFVLLLNM LIALMGETVE NVSKESERIW RLQRARTILE FEKMLPEWLR SRFRMGEL C KVADEDFRLC LRINEVKWTE WKTHVSFLNE DPGPIRRTAD LNKIQDSSRS NSKTTLYAFD ELDEFPETSV LVPRGSAAA WSHPQFEK UniProtKB: Transient receptor potential cation channel subfamily V member 3 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 24 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: Dyclonine
| Macromolecule | Name: Dyclonine / type: ligand / ID: 3 / Number of copies: 4 / Formula: N8R |
|---|---|
| Molecular weight | Theoretical: 289.412 Da |
| Chemical component information | 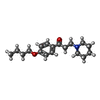 ChemComp-N8R: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 124 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | mouse TRPV3 in circularized NW30 nanodiscs (cNW30) with soybean lipids |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6856 / Average exposure time: 2.5 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)