[English] 日本語
 Yorodumi
Yorodumi- EMDB-26381: Cryo-EM structure of Shiga toxin 2 in complex with the native rib... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Shiga toxin 2 / Ribosomal P-stalk / Complex / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Shigella dysenteriae (bacteria) / Shigella dysenteriae (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Kulczyk AW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction. Authors: Arkadiusz W Kulczyk / Carlos Oscar S Sorzano / Przemysław Grela / Marek Tchórzewski / Nilgun E Tumer / Xiao-Ping Li /    Abstract: Shiga toxin 2a (Stx2a) is the virulence factor of enterohemorrhagic Escherichia coli. The catalytic A1 subunit of Stx2a (Stx2A1) interacts with the ribosomal P-stalk for loading onto the ribosome and ...Shiga toxin 2a (Stx2a) is the virulence factor of enterohemorrhagic Escherichia coli. The catalytic A1 subunit of Stx2a (Stx2A1) interacts with the ribosomal P-stalk for loading onto the ribosome and depurination of the sarcin-ricin loop, which halts protein synthesis. Because of the intrinsic flexibility of the P-stalk, a structure of the Stx2a-P-stalk complex is currently unknown. We demonstrated that the native P-stalk pentamer binds to Stx2a with nanomolar affinity, and we employed cryo-EM to determine a structure of the 72 kDa Stx2a complexed with the P-stalk. The structure identifies Stx2A1 residues involved in binding and reveals that Stx2a is anchored to the P-stalk via only the last six amino acids from the C-terminal domain of a single P-protein. For the first time, the cryo-EM structure shows the loop connecting Stx2A1 and Stx2A2, which is critical for activation of the toxin. Our principal component analysis of the cryo-EM data reveals the intrinsic dynamics of the Stx2a-P-stalk interaction, including conformational changes in the P-stalk binding site occurring upon complex formation. Our computational analysis unveils the propensity for structural rearrangements within the C-terminal domain, with its C-terminal six amino acids transitioning from a random coil to an α-helix upon binding to Stx2a. In conclusion, our cryo-EM structure sheds new light into the dynamics of the Stx2a-P-stalk interaction and indicates that the binding interface between Stx2a and the P-stalk is the potential target for drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26381.map.gz emd_26381.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26381-v30.xml emd-26381-v30.xml emd-26381.xml emd-26381.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26381_fsc.xml emd_26381_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_26381.png emd_26381.png | 61.7 KB | ||
| Filedesc metadata |  emd-26381.cif.gz emd-26381.cif.gz | 5.7 KB | ||
| Others |  emd_26381_half_map_1.map.gz emd_26381_half_map_1.map.gz emd_26381_half_map_2.map.gz emd_26381_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26381 http://ftp.pdbj.org/pub/emdb/structures/EMD-26381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26381 | HTTPS FTP |
-Related structure data
| Related structure data |  7u6vMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26381.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26381.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8248 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26381_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
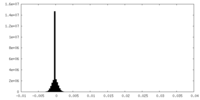
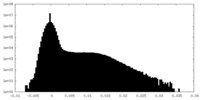
| Density Histograms |
-Half map: #1
| File | emd_26381_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
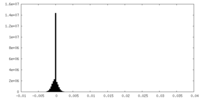
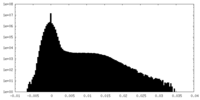
| Density Histograms |
- Sample components
Sample components
-Entire : Shiga toxin 2a in complex with the native ribosomal P-stalk
| Entire | Name: Shiga toxin 2a in complex with the native ribosomal P-stalk |
|---|---|
| Components |
|
-Supramolecule #1: Shiga toxin 2a in complex with the native ribosomal P-stalk
| Supramolecule | Name: Shiga toxin 2a in complex with the native ribosomal P-stalk type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 100 KDa |
-Supramolecule #2: Shiga toxin 2a
| Supramolecule | Name: Shiga toxin 2a / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 / Details: 72 KDa Stx2a holotoxin |
|---|---|
| Source (natural) | Organism:  Shigella dysenteriae (bacteria) Shigella dysenteriae (bacteria) |
-Supramolecule #3: Native ribosomal P-stalk
| Supramolecule | Name: Native ribosomal P-stalk / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 / Details: 56 kDa native ribosomal P-stalk |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Shiga toxin 2a subunit A (Stx2A)
| Macromolecule | Name: Shiga toxin 2a subunit A (Stx2A) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: rRNA N-glycosylase |
|---|---|
| Source (natural) | Organism:  Shigella dysenteriae (bacteria) Shigella dysenteriae (bacteria) |
| Molecular weight | Theoretical: 33.214188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: REFTIDFSTQ QSYVSSLNSI RTEISTPLEH ISQGTTSVSV INHTPPGSYF AVDIRGLDVY QARFDHLRLI IEQNNLYVAG FVNTATNTF YRFSDFTHIS VPGVTTVSMT TDSSYTTLQR VAALERSGMQ ISRHSLVSSY LALMEFSGNT MTRDASRAVL R FVTVTAEA ...String: REFTIDFSTQ QSYVSSLNSI RTEISTPLEH ISQGTTSVSV INHTPPGSYF AVDIRGLDVY QARFDHLRLI IEQNNLYVAG FVNTATNTF YRFSDFTHIS VPGVTTVSMT TDSSYTTLQR VAALERSGMQ ISRHSLVSSY LALMEFSGNT MTRDASRAVL R FVTVTAEA LRFRQIQREF RQALSETAPV YTMTPGDVDL TLNWGRISNV LPEYRGEDGV RVGRISFNNI SAILGTVAVI LN CHHQGAR SVRAVNEDSQ PECQITGDRP VIKINNTLWE SNTAAAFLNR KSQFLYTTGK UniProtKB: rRNA N-glycosylase |
-Macromolecule #2: Shiga toxin 2a subunit B (Stx2B)
| Macromolecule | Name: Shiga toxin 2a subunit B (Stx2B) / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella dysenteriae (bacteria) Shigella dysenteriae (bacteria) |
| Molecular weight | Theoretical: 7.82459 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ADCAKGKIEF SKYNEDDTFT VKVDGKEYWT SRWNLQPLLQ SAQLTGMTVT IKSSTCESGS GFAEVQFNND |
-Macromolecule #3: C-terminal domain (CTD) from the Ribosomal P-stalk
| Macromolecule | Name: C-terminal domain (CTD) from the Ribosomal P-stalk / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 654.712 Da |
| Sequence | String: GFGLFD |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 23 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-7u6v: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)