[English] 日本語
 Yorodumi
Yorodumi- EMDB-26379: Structure of PA28gamma bound to the human 20S proteasome with C7 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PA28gamma bound to the human 20S proteasome with C7 symmetry. | |||||||||
 Map data Map data | Complex of PA28gamma(REGgamma) bound to the human 20S proteasome with C7 symmetry applied. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
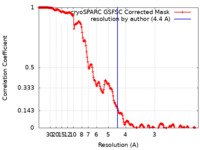
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Smith DM / Thomas T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. Authors: Taylor A Thomas / David M Smith /  Abstract: Proteasome activator 28γ (PA28γ/REGγ) is a member of the 11S family of proteasomal regulators that is constitutively expressed in the nucleus and implicated in various diseases, including certain ...Proteasome activator 28γ (PA28γ/REGγ) is a member of the 11S family of proteasomal regulators that is constitutively expressed in the nucleus and implicated in various diseases, including certain cancers and systemic lupus erythematosus. Despite years of investigation, how PA28γ functions to stimulate proteasomal protein degradation remains unclear. Alternative hypotheses have been proposed for the molecular mechanism of PA28γ, including the following: (1) substrate selection, (2) allosteric upregulation of the trypsin-like (T-L) site, (3) allosteric inhibition of the chymotrypsin-like (CT-L) and caspase-like (C-L) sites, (4) conversion of the CT-L or C-L sites to new T-L sites, and (5) gate opening alone or in combination with a previous hypothesis. Here, by mechanistically decoupling gating effects from active site effects, we unambiguously demonstrate that WT PA28γ allosterically activates the T-L site. We show PA28γ binding increases the Kcat/Km by 13-fold for T-L peptide substrates while having little-to-no effect on hydrolysis kinetics for CT-L or C-L substrates. Furthermore, mutagenesis and domain swaps of PA28γ reveal that it does not select for T-L peptide substrates through either the substrate entry pore or the distal intrinsically disordered region. We also show that a previously reported point mutation can functionally switch PA28γ from a T-L activating to a gate-opening activator in a mutually exclusive fashion. Finally, using cryogenic electron microscopy, we visualized the PA28γ-proteasome complex at 4.3 Å and confirmed its expected quaternary structure. The results of this study provide unambiguous evidence that PA28γ can function by binding the 20S proteasome to allosterically activate the T-L proteolytic site. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26379.map.gz emd_26379.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26379-v30.xml emd-26379-v30.xml emd-26379.xml emd-26379.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26379_fsc.xml emd_26379_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_26379.png emd_26379.png | 91.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26379 http://ftp.pdbj.org/pub/emdb/structures/EMD-26379 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26379 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26379 | HTTPS FTP |
-Validation report
| Summary document |  emd_26379_validation.pdf.gz emd_26379_validation.pdf.gz | 563.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26379_full_validation.pdf.gz emd_26379_full_validation.pdf.gz | 563 KB | Display | |
| Data in XML |  emd_26379_validation.xml.gz emd_26379_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_26379_validation.cif.gz emd_26379_validation.cif.gz | 22.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26379 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26379 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26379 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26379 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26379.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26379.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex of PA28gamma(REGgamma) bound to the human 20S proteasome with C7 symmetry applied. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : CryoEM structure of the PA28gamma 20S proteasome complex.
| Entire | Name: CryoEM structure of the PA28gamma 20S proteasome complex. |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of the PA28gamma 20S proteasome complex.
| Supramolecule | Name: CryoEM structure of the PA28gamma 20S proteasome complex. type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all / Details: C7 symmetry is applied. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 900 KDa |
-Macromolecule #1: PA28 gamma
| Macromolecule | Name: PA28 gamma / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MASLLKVDQE VKLKVDSFRE RITSEAEDLV ANFFPKKLLE LDSFLKEPIL NIHDLTQIHS DMNLPVPDPI LLTNSHDGLD GPTYKKRRLD ECEEAFQGTK VFVMPNGMLK SNQQLVDIIE KVKPEIRLLI EKCNTVKMWV QLLIPRIEDG NNFGVSIQEE TVAELRTVES ...String: MASLLKVDQE VKLKVDSFRE RITSEAEDLV ANFFPKKLLE LDSFLKEPIL NIHDLTQIHS DMNLPVPDPI LLTNSHDGLD GPTYKKRRLD ECEEAFQGTK VFVMPNGMLK SNQQLVDIIE KVKPEIRLLI EKCNTVKMWV QLLIPRIEDG NNFGVSIQEE TVAELRTVES EAASYLDQIS RYYITRAKLV SKIAKYPHVE DYRRTVTEID EKEYISLRLI ISELRNQYVT LHDMILKNIE KIKRPRSSNA ETLY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 50.0 mM / Component - Formula: Tris-HCL / Component - Name: Tris buffer |
| Grid | Model: Quantifoil / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Details | Recombinate human PA28gamma was combine with human 20S proteasome from Hek293 cells. The sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)