[English] 日本語
 Yorodumi
Yorodumi- EMDB-26344: YcaO-mediated ATP-dependent peptidase activity in ribosomal pepti... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | YcaO-mediated ATP-dependent peptidase activity in ribosomal peptide biosynthesis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ripps / Ycao domain / musD / Cyanobactin / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Desmonostoc sp. PCC 7906 (bacteria) Desmonostoc sp. PCC 7906 (bacteria) | |||||||||
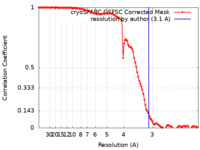
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zheng Y / Nair SK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: YcaO-mediated ATP-dependent peptidase activity in ribosomal peptide biosynthesis. Authors: Yiwu Zheng / Satish K Nair /  Abstract: YcaO enzymes catalyze ATP-dependent post-translation modifications on peptides, including the installation of (ox/thi)azoline, thioamide and/or amidine moieties. Here we demonstrate that, in the ...YcaO enzymes catalyze ATP-dependent post-translation modifications on peptides, including the installation of (ox/thi)azoline, thioamide and/or amidine moieties. Here we demonstrate that, in the biosynthesis of the bis-methyloxazolic alkaloid muscoride A, the YcaO enzyme MusD carries out both ATP-dependent cyclodehydration and peptide bond cleavage, which is a mechanism unprecedented for such a reaction. YcaO-catalyzed modifications are proposed to occur through a backbone O-phosphorylated intermediate, but this mechanism remains speculative. We report, to our knowedge, the first characterization of an acyl-phosphate species consistent with the proposed mechanism for backbone amide activation. The 3.1-Å-resolution cryogenic electron microscopy structure of MusD along with biochemical analysis allow identification of residues that enable peptide cleavage reaction. Bioinformatics analysis identifies other cyanobactin pathways that may deploy bifunctional YcaO enzymes. Our structural, mutational and mechanistic studies expand the scope of modifications catalyzed by YcaO proteins to include peptide hydrolysis and provide evidence for a unifying mechanism for the catalytically diverse outcomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26344.map.gz emd_26344.map.gz | 62.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26344-v30.xml emd-26344-v30.xml emd-26344.xml emd-26344.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26344_fsc.xml emd_26344_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_26344.png emd_26344.png | 104.4 KB | ||
| Masks |  emd_26344_msk_1.map emd_26344_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26344.cif.gz emd-26344.cif.gz | 5.7 KB | ||
| Others |  emd_26344_half_map_1.map.gz emd_26344_half_map_1.map.gz emd_26344_half_map_2.map.gz emd_26344_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26344 http://ftp.pdbj.org/pub/emdb/structures/EMD-26344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26344 | HTTPS FTP |
-Related structure data
| Related structure data |  7u58MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26344.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26344.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26344_msk_1.map emd_26344_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26344_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26344_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MusD
| Entire | Name: MusD |
|---|---|
| Components |
|
-Supramolecule #1: MusD
| Supramolecule | Name: MusD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Desmonostoc sp. PCC 7906 (bacteria) Desmonostoc sp. PCC 7906 (bacteria) |
| Molecular weight | Theoretical: 88.76 kDa/nm |
-Macromolecule #1: MusD
| Macromolecule | Name: MusD / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desmonostoc sp. PCC 7906 (bacteria) / Strain: PCC 7906 Desmonostoc sp. PCC 7906 (bacteria) / Strain: PCC 7906 |
| Molecular weight | Theoretical: 88.844805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMKILQIK PHFRAEIIEP KHVYLLSESS THALTGELYC QLIPLLNGNY TVDEIINKLQ VDPSHIDYAL ERLQARGYIT EAIPQLTPE AVAFWGLLKV EPQVAYQCLQ QTQVYVSSVV NLPTQPLITA LEEVGIKAIN WDGELQEFPP HSLLVVLTDD Y LQPQLNKI ...String: SNAMKILQIK PHFRAEIIEP KHVYLLSESS THALTGELYC QLIPLLNGNY TVDEIINKLQ VDPSHIDYAL ERLQARGYIT EAIPQLTPE AVAFWGLLKV EPQVAYQCLQ QTQVYVSSVV NLPTQPLITA LEEVGIKAIN WDGELQEFPP HSLLVVLTDD Y LQPQLNKI NQIALKANQP WLLIKPVGTI LWLGPIFQPQ ITGCWECLAQ RLRVNREVEA SVLRQKNSSL QLSPSQELNS SV LQSNGNG VKSEVIECLP PPAAVIPSTL QTALHLATTE IAKWIVKQGV EDTTPFPTLE GKVITFDQRN LDLQTHILSL RPQ CPSCGN PNLLTERAFQ PLVLSSRKKQ FTSDGGHRAF SPDQTVNRYQ HLISPITGVV TSLVRASDPN DSLNHTYNAV HSFV IASNI GRMRRYLKHK SSGKGKTDSQ SKASGFCEAI ERYSGVYQGD EPRISATLAE LGEKAIHPAR CSLFSSEQYE YREEF NRRG GVFDWIPQPF DETKVIEWTP VWSLTEQTHK YIPTAYCYYG YPLPEDHEFC RANSNGDATG NTLEEAIIQG FFEIVE RDS VAIWWYNRLK RPAVDLASFN EPYLLEVQDL YRSNNRDLWV IDITADLDIP TFVAVSYLKD NKHQTILLGF GTHFDPK IA ILRAVTEVNQ IAFTCDGVEV TKEFVEMREW FKKATIENQP YLVPDSTVPA KVYQDYQQRW SDDIYEDVMT CVEISKNA G LETLVLDKTR PDIGLNVAKV IVPEMPHYWL RMGAKRIYDV PVKMGWLSTP LTEEQMNPIS VPI UniProtKB: MusD |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: HEPES pH 7.5. 25 mM KCl. |
| Grid | Material: COPPER |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.33 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)