[English] 日本語
 Yorodumi
Yorodumi- EMDB-26161: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q with ATP and 16x(Asp-Arg) | |||||||||
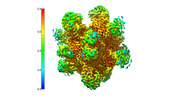
 Map data Map data | Locally filtered map used for refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cyanophycin / CphA1 / ATP-grasp / enzyme / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcyanophycin synthase (L-aspartate-adding) / cyanophycin synthase (L-arginine-adding) / cyanophycin synthetase activity (L-aspartate-adding) / cyanophycin synthetase activity (L-arginine-adding) / biosynthetic process / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
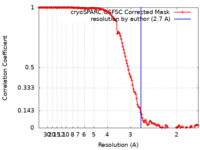
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Sharon I / Grogg M | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: A cryptic third active site in cyanophycin synthetase creates primers for polymerization Authors: Sharon I / Pinus S / Grogg M / Moitessier N / Hilvert D / Schmeing TM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26161.map.gz emd_26161.map.gz | 11 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26161-v30.xml emd-26161-v30.xml emd-26161.xml emd-26161.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26161_fsc.xml emd_26161_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26161.png emd_26161.png | 51.8 KB | ||
| Masks |  emd_26161_msk_1.map emd_26161_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26161.cif.gz emd-26161.cif.gz | 6.1 KB | ||
| Others |  emd_26161_additional_1.map.gz emd_26161_additional_1.map.gz emd_26161_half_map_1.map.gz emd_26161_half_map_1.map.gz emd_26161_half_map_2.map.gz emd_26161_half_map_2.map.gz | 230 MB 226.1 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26161 http://ftp.pdbj.org/pub/emdb/structures/EMD-26161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26161 | HTTPS FTP |
-Validation report
| Summary document |  emd_26161_validation.pdf.gz emd_26161_validation.pdf.gz | 892.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26161_full_validation.pdf.gz emd_26161_full_validation.pdf.gz | 892.3 KB | Display | |
| Data in XML |  emd_26161_validation.xml.gz emd_26161_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_26161_validation.cif.gz emd_26161_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26161 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26161 | HTTPS FTP |
-Related structure data
| Related structure data |  7txvMC  7txuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26161.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26161.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map used for refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
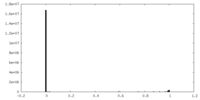
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26161_msk_1.map emd_26161_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
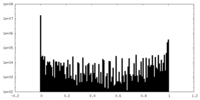
| Density Histograms |
-Additional map: Sharpened map before local filtering. Not used for refinement.
| File | emd_26161_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map before local filtering. Not used for refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
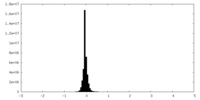
| Density Histograms |
-Half map: half map A
| File | emd_26161_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
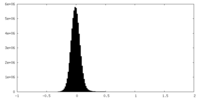
| Density Histograms |
-Half map: half map B
| File | emd_26161_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q wit...
| Entire | Name: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q with ATP and 16x(Asp-Arg) |
|---|---|
| Components |
|
-Supramolecule #1: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q wit...
| Supramolecule | Name: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q with ATP and 16x(Asp-Arg) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 383 kDa/nm |
-Macromolecule #1: Cyanophycin synthase
| Macromolecule | Name: Cyanophycin synthase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: cyanophycin synthase (L-aspartate-adding) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.757852 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKILKTLTLR GPNYWSIRRK KLIVMRLDLE DLAERPSNSI PGFYEGLIKV LPSLVEHFCS PGYQGGFLER VKEGTYMGHI VQHVALELQ ELVGMTAGFG RTRETSTPGV YNVVYEYVDE QAGRYAGRAA VRLCRSLVDT GDYPRLELEK DLEDLRDLGA N SALGPSTE ...String: MKILKTLTLR GPNYWSIRRK KLIVMRLDLE DLAERPSNSI PGFYEGLIKV LPSLVEHFCS PGYQGGFLER VKEGTYMGHI VQHVALELQ ELVGMTAGFG RTRETSTPGV YNVVYEYVDE QAGRYAGRAA VRLCRSLVDT GDYPRLELEK DLEDLRDLGA N SALGPSTE TIVTEAEARK IPWMLLSARA MVQLGYGVYQ QRIQATLSSH SGILGVELAC DKEGTKTILQ DAGIPVPRGT TI QYFDDLE EAINDVGGYP VVIKPLDGNH GRGITINVRH WQEAIAAYDL AAEESKSRAI IVERYYEGSD HRVLVVNGKL VAV AERIPA HVTGDGSSTI SELIEKTNQD PNRGDGHDNI LTKIVVNKTA IDVMERQGYN LDSVLPKDEV VYLRATANLS TGGI AIDRT DDIHPENIWL MERVAKVIGL DIAGIDVVTS DISKPLRETN GVIVEVNAAP GFRMHVAPSQ GLPRNVAAPV LDMLF PPGT PSRIPILAVT GTNGKTTTTR LLAHIYRQTG KTVGYTSTDA IYINEYCVEK GDNTGPQSAG VILRDPTVEV AVLETA RGG ILRAGLAFDS CDVGVVLNVA ADHLGLGDID TIEQMAKVKS VIAEVVDPSG YAVLNADDPL VAAMADKVKA KVAYFSM NP DNPIIQAHVR RNGIAAVYES GYLSILEGSW TLRVEQAKLI PMTMGGMAPF MIANALAACL AAFVNGLDVE VIRQGVRT F TTSAEQTPGR MNLFNLGQHH ALVDYAHNPA GYRAVGDFVK NWQGQRFGVV GGPGDRRDSD LIELGQIAAQ VFDRIIVKE DDDKRGRSEG ETADLIVKGI LQENPGASYE VILDETIALN KALDQVEEKG LVVVFPESVT RAIDLIKVRN PIGENLYFQ UniProtKB: Cyanophycin synthetase |
-Macromolecule #2: 16x(Asp-Arg)
| Macromolecule | Name: 16x(Asp-Arg) / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.35838 KDa |
| Sequence | String: (7ID)(7ID)(7ID)(7ID)(7ID)(7ID)(7ID)(7ID)(7ID)(7ID) (7ID)(7ID)(7ID)(7ID)(7ID)(7ID) |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 16 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 8 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)