[English] 日本語
 Yorodumi
Yorodumi- EMDB-26101: Human M4 muscarinic acetylcholine receptor complex with Gi1 and t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo and positive allosteric modulator VU0467154 | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | 7 transmembrane receptor / SIGNALING PROTEIN-IMMUNE SYSTEM complex | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMuscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of locomotion / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E ...Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of locomotion / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Ras protein signal transduction / postsynaptic membrane / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / dendrite / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
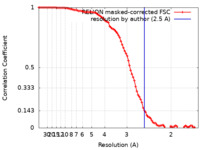
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||||||||||||||
 Authors Authors | Vuckovic Z / Mobbs JI | ||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Australia, Australia,  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor Authors: Thal DM | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26101.map.gz emd_26101.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26101-v30.xml emd-26101-v30.xml emd-26101.xml emd-26101.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26101_fsc.xml emd_26101_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26101.png emd_26101.png | 111.1 KB | ||
| Masks |  emd_26101_msk_1.map emd_26101_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26101.cif.gz emd-26101.cif.gz | 7.2 KB | ||
| Others |  emd_26101_half_map_1.map.gz emd_26101_half_map_1.map.gz emd_26101_half_map_2.map.gz emd_26101_half_map_2.map.gz | 49.8 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26101 http://ftp.pdbj.org/pub/emdb/structures/EMD-26101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26101 | HTTPS FTP |
-Related structure data
| Related structure data |  7trqMC  7trkC  7trpC  7trsC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26101.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26101.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26101_msk_1.map emd_26101_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26101_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26101_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : M4 mAChR bound to agonist iperoxo and the PAM VU0467154 in comple...
| Entire | Name: M4 mAChR bound to agonist iperoxo and the PAM VU0467154 in complex with dominant negative Galpha-i1, Gbeta1, Ggamma2, and scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: M4 mAChR bound to agonist iperoxo and the PAM VU0467154 in comple...
| Supramolecule | Name: M4 mAChR bound to agonist iperoxo and the PAM VU0467154 in complex with dominant negative Galpha-i1, Gbeta1, Ggamma2, and scFv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Muscarinic acetylcholine receptor M4
| Macromolecule | Name: Muscarinic acetylcholine receptor M4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.598277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAMA NFTPVNGSSG NQSVRLVTSS SHNRYETVEM VFIATVTGSL SLVTVVGNIL VMLSIKVNRQ LQTVNNYFLF SLACADLII GAFSMNLYTV YIIKGYWPLG AVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP ARRTTKMAGL M IAAAWVLS ...String: DYKDDDDAMA NFTPVNGSSG NQSVRLVTSS SHNRYETVEM VFIATVTGSL SLVTVVGNIL VMLSIKVNRQ LQTVNNYFLF SLACADLII GAFSMNLYTV YIIKGYWPLG AVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP ARRTTKMAGL M IAAAWVLS FVLWAPAILF WQFVVGKRTV PDNQCFIQFL SNPAVTFGTA IAAFYLPVVI MTVLYIHISL ASRSRVHKHR PE GPKEKKA KTKRQMAARE RKVTRTIFAI LLAFILTWTP YNVMVLVNTF CQSCIPDTVW SIGYWLCYVN STINPACYAL CNA TFKKTF RHLLLCQYRN IGTARHHHHH HHH UniProtKB: Muscarinic acetylcholine receptor M4, Muscarinic acetylcholine receptor M4 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.402867 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: HHHHHHGSSG SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLI IWDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR F LDDNQIVT ...String: HHHHHHGSSG SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLI IWDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR F LDDNQIVT SSGDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NA ICFFPNG NAFATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGH DNRVSC LGVTDDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Antibody fragment scFv16
| Macromolecule | Name: Antibody fragment scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.466486 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL K |
-Macromolecule #5: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #6: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium
| Macromolecule | Name: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium type: ligand / ID: 6 / Number of copies: 1 / Formula: IXO |
|---|---|
| Molecular weight | Theoretical: 197.254 Da |
| Chemical component information | 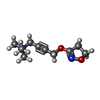 ChemComp-IXO: |
-Macromolecule #7: 5-amino-3,4-dimethyl-N-{[4-(trifluoromethanesulfonyl)phenyl]methy...
| Macromolecule | Name: 5-amino-3,4-dimethyl-N-{[4-(trifluoromethanesulfonyl)phenyl]methyl}thieno[2,3-c]pyridazine-6-carboxamide type: ligand / ID: 7 / Number of copies: 1 / Formula: IUI |
|---|---|
| Molecular weight | Theoretical: 444.451 Da |
| Chemical component information | 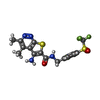 ChemComp-IUI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)