+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
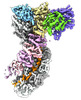
| タイトル | Cascade complex from type I-A CRISPR-Cas system | ||||||||||||
 マップデータ マップデータ | |||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | CRISPR / cascade / Cas3 / type I-A / genome editing / RNA BINDING PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報catalytic activity, acting on DNA / exonuclease activity / endonuclease activity / defense response to virus / nucleic acid binding / 加水分解酵素; エステル加水分解酵素 / RNA helicase activity / ATP binding / metal ion binding / cytosol 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  Geobacter sulfurreducens (バクテリア) / Geobacter sulfurreducens (バクテリア) /   Pyrococcus furiosus DSM 3638 (古細菌) / Pyrococcus furiosus DSM 3638 (古細菌) /  | ||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.6 Å | ||||||||||||
 データ登録者 データ登録者 | Hu C / Ni D | ||||||||||||
| 資金援助 |  米国, 米国,  スイス, 3件 スイス, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2022 ジャーナル: Mol Cell / 年: 2022タイトル: Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools. 著者: Chunyi Hu / Dongchun Ni / Ki Hyun Nam / Sonali Majumdar / Justin McLean / Henning Stahlberg / Michael P Terns / Ailong Ke /    要旨: Type I CRISPR-Cas systems typically rely on a two-step process to degrade DNA. First, an RNA-guided complex named Cascade identifies the complementary DNA target. The helicase-nuclease fusion enzyme ...Type I CRISPR-Cas systems typically rely on a two-step process to degrade DNA. First, an RNA-guided complex named Cascade identifies the complementary DNA target. The helicase-nuclease fusion enzyme Cas3 is then recruited in trans for processive DNA degradation. Contrary to this model, here, we show that type I-A Cascade and Cas3 function as an integral effector complex. We provide four cryoelectron microscopy (cryo-EM) snapshots of the Pyrococcus furiosus (Pfu) type I-A effector complex in different stages of DNA recognition and degradation. The HD nuclease of Cas3 is autoinhibited inside the effector complex. It is only allosterically activated upon full R-loop formation, when the entire targeted region has been validated by the RNA guide. The mechanistic insights inspired us to convert Pfu Cascade-Cas3 into a high-sensitivity, low-background, and temperature-activated nucleic acid detection tool. Moreover, Pfu CRISPR-Cas3 shows robust bi-directional deletion-editing activity in human cells, which could find usage in allele-specific inactivation of disease-causing mutations. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_26082.map.gz emd_26082.map.gz | 32.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-26082-v30.xml emd-26082-v30.xml emd-26082.xml emd-26082.xml | 25.8 KB 25.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_26082.png emd_26082.png | 143.5 KB | ||
| Filedesc metadata |  emd-26082.cif.gz emd-26082.cif.gz | 8.1 KB | ||
| その他 |  emd_26082_additional_1.map.gz emd_26082_additional_1.map.gz | 59.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26082 http://ftp.pdbj.org/pub/emdb/structures/EMD-26082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26082 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_26082.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_26082.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.6336 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: #1
| ファイル | emd_26082_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Cas3 bound Cascade complex from Pyrococcus furiosus type I-A CRIS...
| 全体 | 名称: Cas3 bound Cascade complex from Pyrococcus furiosus type I-A CRISPR/Cas system |
|---|---|
| 要素 |
|
-超分子 #1: Cas3 bound Cascade complex from Pyrococcus furiosus type I-A CRIS...
| 超分子 | 名称: Cas3 bound Cascade complex from Pyrococcus furiosus type I-A CRISPR/Cas system タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#7 / 詳細: Cascade alone |
|---|---|
| 由来(天然) | 生物種:  Geobacter sulfurreducens (バクテリア) Geobacter sulfurreducens (バクテリア) |
| 分子量 | 理論値: 400 KDa |
-分子 #1: CRISPR-associated helicase Cas3
| 分子 | 名称: CRISPR-associated helicase Cas3 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 61.412555 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: ELTGFEPYDY QLRAWEKIRE IMNNGGKVII EVPTAGGKTE TAVMPFFAGI YNNNWPVARL VYVLPTRSLV EKQAERLRNL VYKLLQLKG KSKEEAEKLA RELVVVEYGL EKTHAFLGWV VVTTWDAFLY GLAAHRTVGN RFTFPAGAIA QSLVIFDEVQ M YQDESMYM ...文字列: ELTGFEPYDY QLRAWEKIRE IMNNGGKVII EVPTAGGKTE TAVMPFFAGI YNNNWPVARL VYVLPTRSLV EKQAERLRNL VYKLLQLKG KSKEEAEKLA RELVVVEYGL EKTHAFLGWV VVTTWDAFLY GLAAHRTVGN RFTFPAGAIA QSLVIFDEVQ M YQDESMYM PRLLSLVVGI LEEANVPLVI MSATIPSKLR EMIAGDTEVI TVDKNDKNKP SKGNVKVRLV EGDITDVLND IK KILKNGK KVLVVRNTVR KAVETYQVLK KKLNDTLANP SDALLIHSRF TIGDRREKER ALDSARLIVA TQVVEAGLDL PNV GLVVTD IAPLDALIQR IGRCARRPGE EGEGIILIPA AAAAAAAAAA AAAAAAAAAA AAAAAAAVVT STNEYDRVVE IHYG EGKKN FVYVGDIDTA RRVLEKKRSK KLPKDLYIIP YSVSPYPDPL VLLTTYDELS KIGEYLADTT KARKALDRVY KFHYE NNIV PKEFASYIYF KELKLFSAPP EYEKAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAI DAKYYNSELA AAAAAA AAA AAAAAAAAA UniProtKB: CRISPR-associated helicase Cas3 |
-分子 #2: Cas8
| 分子 | 名称: Cas8 / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 38.800719 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: FNEFKTPQID PIFDLYVAYG YVVSLIRGGA KEATLIPHGA SYLIQTDVSN EEFRHGLVDA LSSMLSLHIA LARHSPREGG KLVSDADFS AGANINNVYW DSVPRNLEKL MKDLEKKRSV KGTATIPITL MPSAGKYMLK HFGVQGGNPI KVDLLNYALA W VGFHYYTP ...文字列: FNEFKTPQID PIFDLYVAYG YVVSLIRGGA KEATLIPHGA SYLIQTDVSN EEFRHGLVDA LSSMLSLHIA LARHSPREGG KLVSDADFS AGANINNVYW DSVPRNLEKL MKDLEKKRSV KGTATIPITL MPSAGKYMLK HFGVQGGNPI KVDLLNYALA W VGFHYYTP YIKYAKGDTT WIHIYQIAPV EEVDMISILS LKDLKMHLPH YYESNLDFLI NRRLALLYHL LHSEALELFT EK EFVIHSY TLERSGNNQA IRSFEEEEIG KLMDFLWKLK RRDFYHAIKF IDDLLKKATE GALALIDAIM NERLEGFYTA LKL GKKAGV VSSREIVAAL EDIIC UniProtKB: Type I-A CRISPR-associated protein Cas8a2/Csa4 |
-分子 #3: Cas11a
| 分子 | 名称: Cas11a / タイプ: protein_or_peptide / ID: 3 / コピー数: 5 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 12.208933 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GGWIRNIGRY LSYLVDDTFE EYAYDVVDGI AKARTQEELL EGVYKALRLA PKLKKKAESK GCPPPRIPSP EDIEALEEKV EQLSNPKDL RKLAVSLALW AFASWNNCP UniProtKB: Uncharacterized protein |
-分子 #4: Cas7a
| 分子 | 名称: Cas7a / タイプ: protein_or_peptide / ID: 4 / コピー数: 7 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 36.989148 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MYVRISGRIR LNAHSLNAQG GGGTNYIEIT KTKVTVRTEN GWTVVEVPAI TGNMLKHWHF VGFVDYFKTT PYGVNLTERA LRYNGTRFG QGETTATKAN GATVQLNDEA TIIKELADAD VHGFLAPKTG RRRVSLVKAS FILPTEDFIK EVEGERLITA I KHNRVDVD ...文字列: MYVRISGRIR LNAHSLNAQG GGGTNYIEIT KTKVTVRTEN GWTVVEVPAI TGNMLKHWHF VGFVDYFKTT PYGVNLTERA LRYNGTRFG QGETTATKAN GATVQLNDEA TIIKELADAD VHGFLAPKTG RRRVSLVKAS FILPTEDFIK EVEGERLITA I KHNRVDVD EKGAIGSSKE GTAQMLFSRE YATGLYGFSI VLDLGLVGIP QGLPVKFEEN QPRPNIVIDP NERKARIESA LK ALIPMLS GYIGANLARS FPVFKVEELV AIASEGPIPA LVHGFYEDYI EANRSIIKNA RALGFNIEVF TYNVDLGEDI EAT KVSSVE ELVANLVKMV UniProtKB: Type I-A CRISPR-associated protein Cas7/Csa2 |
-分子 #5: Cas5a
| 分子 | 名称: Cas5a / タイプ: protein_or_peptide / ID: 5 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 29.147219 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MDILLVCLRF PFFSVAKRSY QVRTSFLLPP PSALKGALAK GLILLKPEKY ASSSLDEAAL KAIKEIESKL VDIKAVSVAP LSPLIRNAF LLKRLRNLES GSNAEKSDAM RREYTFTREL LVAYIFKNLT QEEKNLYLKA AMLIDVIGDT ESLATPVWAS F VKPEDKKA ...文字列: MDILLVCLRF PFFSVAKRSY QVRTSFLLPP PSALKGALAK GLILLKPEKY ASSSLDEAAL KAIKEIESKL VDIKAVSVAP LSPLIRNAF LLKRLRNLES GSNAEKSDAM RREYTFTREL LVAYIFKNLT QEEKNLYLKA AMLIDVIGDT ESLATPVWAS F VKPEDKKA PLAFSAPYTE IYSLLSSKIQ AKGKIRMYIE KMRVSPEYSK TKGPQEEIFY LPIEERRYKR IVYYARIYPP EV EKALTVD GEVLGIWIP UniProtKB: Type I-A CRISPR-associated protein Cas5 |
-分子 #6: CRISPR-associated endonuclease Cas3-HD
| 分子 | 名称: CRISPR-associated endonuclease Cas3-HD / タイプ: protein_or_peptide / ID: 6 / コピー数: 1 / 光学異性体: LEVO EC番号: 加水分解酵素; エステル加水分解酵素 |
|---|---|
| 由来(天然) | 生物種:   Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus DSM 3638 (古細菌) / 株: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| 分子量 | 理論値: 26.057617 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: SCKAFQGQTL REHIEAMLAA WEIVKNKYIP SIIRVMKTVG VKFTEEDADK FMKTLIILHD VGKCSEVYQK HLSNNEPLRG FRHELVSAY YAYNILKDMF KDETIAFIGA LVVMMHHEPI LMGQIRSLDK EELTPEVVLD KLRTFNGVME GTESFIKSMI K EKLGVIPK ...文字列: SCKAFQGQTL REHIEAMLAA WEIVKNKYIP SIIRVMKTVG VKFTEEDADK FMKTLIILHD VGKCSEVYQK HLSNNEPLRG FRHELVSAY YAYNILKDMF KDETIAFIGA LVVMMHHEPI LMGQIRSLDK EELTPEVVLD KLRTFNGVME GTESFIKSMI K EKLGVIPK VPSPTQEDVL REVIRLSVLA RHRPDSGKLR MVVGALLIPL VCDYKGAAAA AAAAAAAAAA AAAAAAAAA UniProtKB: CRISPR-associated endonuclease Cas3-HD |
-分子 #7: crRNA (45-MER)
| 分子 | 名称: crRNA (45-MER) / タイプ: rna / ID: 7 / コピー数: 1 |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 14.373537 KDa |
| 配列 | 文字列: AUUGAAAGAG UGCUUCCCCA AACCCUUAAC UGGUUGUAAC AGUUG |
-分子 #8: NICKEL (II) ION
| 分子 | 名称: NICKEL (II) ION / タイプ: ligand / ID: 8 / コピー数: 2 / 式: NI |
|---|---|
| 分子量 | 理論値: 58.693 Da |
| Chemical component information |  ChemComp-NI: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 / 構成要素 - 濃度: 150.0 mM / 構成要素 - 式: NaCl / 構成要素 - 名称: sodium chloride |
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 前処理 - タイプ: PLASMA CLEANING / 前処理 - 時間: 30 sec. |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 298 K / 装置: FEI VITROBOT MARK IV / 詳細: 6 seconds. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS TALOS |
|---|---|
| 特殊光学系 | 位相板: VOLTA PHASE PLATE |
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 実像数: 822 / 平均露光時間: 2.5 sec. / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 100.0 µm / 最小 デフォーカス(補正後): 1.0 µm / 照射モード: FLOOD BEAM / 撮影モード: DIFFRACTION / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.5 µm |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | プロトコル: OTHER |
|---|---|
| 得られたモデル |  PDB-7tr8: |
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




























