[English] 日本語
 Yorodumi
Yorodumi- EMDB-26054: Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Lipopolysaccharide / single-particle cryo-electron microscopy / molecular dynamics simulations / structural biology / undecaprenyl pyrophosphate / WaaL Ligase / lipid A / O-antigen / membrane proteins / transglycosylation / glycosyltransferase / MEMBRANE PROTEIN | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology | O-antigen ligase-related / : / O-Antigen ligase / membrane / Cell surface polysaccharide polymerase/ligase Function and homology information Function and homology information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Cupriavidus metallidurans (bacteria) / Cupriavidus metallidurans (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Ashraf KU / Nygaard R | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, 18 items United Kingdom, 18 items
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural basis of lipopolysaccharide maturation by the O-antigen ligase. Authors: Khuram U Ashraf / Rie Nygaard / Owen N Vickery / Satchal K Erramilli / Carmen M Herrera / Thomas H McConville / Vasileios I Petrou / Sabrina I Giacometti / Meagan Belcher Dufrisne / Kamil ...Authors: Khuram U Ashraf / Rie Nygaard / Owen N Vickery / Satchal K Erramilli / Carmen M Herrera / Thomas H McConville / Vasileios I Petrou / Sabrina I Giacometti / Meagan Belcher Dufrisne / Kamil Nosol / Allen P Zinkle / Chris L B Graham / Michael Loukeris / Brian Kloss / Karolina Skorupinska-Tudek / Ewa Swiezewska / David I Roper / Oliver B Clarke / Anne-Catrin Uhlemann / Anthony A Kossiakoff / M Stephen Trent / Phillip J Stansfeld / Filippo Mancia /    Abstract: The outer membrane of Gram-negative bacteria has an external leaflet that is largely composed of lipopolysaccharide, which provides a selective permeation barrier, particularly against antimicrobials. ...The outer membrane of Gram-negative bacteria has an external leaflet that is largely composed of lipopolysaccharide, which provides a selective permeation barrier, particularly against antimicrobials. The final and crucial step in the biosynthesis of lipopolysaccharide is the addition of a species-dependent O-antigen to the lipid A core oligosaccharide, which is catalysed by the O-antigen ligase WaaL. Here we present structures of WaaL from Cupriavidus metallidurans, both in the apo state and in complex with its lipid carrier undecaprenyl pyrophosphate, determined by single-particle cryo-electron microscopy. The structures reveal that WaaL comprises 12 transmembrane helices and a predominantly α-helical periplasmic region, which we show contains many of the conserved residues that are required for catalysis. We observe a conserved fold within the GT-C family of glycosyltransferases and hypothesize that they have a common mechanism for shuttling the undecaprenyl-based carrier to and from the active site. The structures, combined with genetic, biochemical, bioinformatics and molecular dynamics simulation experiments, offer molecular details on how the ligands come in apposition, and allows us to propose a mechanistic model for catalysis. Together, our work provides a structural basis for lipopolysaccharide maturation in a member of the GT-C superfamily of glycosyltransferases. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26054.map.gz emd_26054.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26054-v30.xml emd-26054-v30.xml emd-26054.xml emd-26054.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26054.png emd_26054.png | 74.1 KB | ||
| Filedesc metadata |  emd-26054.cif.gz emd-26054.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26054 http://ftp.pdbj.org/pub/emdb/structures/EMD-26054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26054 | HTTPS FTP |
-Validation report
| Summary document |  emd_26054_validation.pdf.gz emd_26054_validation.pdf.gz | 567.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26054_full_validation.pdf.gz emd_26054_full_validation.pdf.gz | 567.1 KB | Display | |
| Data in XML |  emd_26054_validation.xml.gz emd_26054_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_26054_validation.cif.gz emd_26054_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26054 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26054 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26054 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26054 | HTTPS FTP |
-Related structure data
| Related structure data |  7tpgMC  7tpjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10938 (Title: CryoEM Structure of the, UND-PP bound, WaaL O-Antigen Ligase EMPIAR-10938 (Title: CryoEM Structure of the, UND-PP bound, WaaL O-Antigen LigaseData size: 477.2 Data #1: Unaligned multi-frame micrographs of the WaaL O-antigen Ligase from cupriavidus metallidurans [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26054.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26054.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Ligand Bound CmWaaL with Fab fragment bound
| Entire | Name: Ligand Bound CmWaaL with Fab fragment bound |
|---|---|
| Components |
|
-Supramolecule #1: Ligand Bound CmWaaL with Fab fragment bound
| Supramolecule | Name: Ligand Bound CmWaaL with Fab fragment bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Macromolecule #1: Putative cell surface polysaccharide polymerase/ligase
| Macromolecule | Name: Putative cell surface polysaccharide polymerase/ligase type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cupriavidus metallidurans (bacteria) Cupriavidus metallidurans (bacteria) |
| Molecular weight | Theoretical: 44.536754 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDSNKLLSR MATVLVFAFP VLILCVPRGA GVFLAGVGVL ALLGWRGMGR AWREYSKVMT PLAIAVLAFM LVYVGSKLYF HTPWNVIDN PSRTLLAILT CWVIVRAAPN PAWLWRGITV GLFLALLIVG YQKFALNIDR PSAWIQAIAF ANMIAALALV G FARPGDSR ...String: MTDSNKLLSR MATVLVFAFP VLILCVPRGA GVFLAGVGVL ALLGWRGMGR AWREYSKVMT PLAIAVLAFM LVYVGSKLYF HTPWNVIDN PSRTLLAILT CWVIVRAAPN PAWLWRGITV GLFLALLIVG YQKFALNIDR PSAWIQAIAF ANMIAALALV G FARPGDSR GTHMEAWVNL LLGTMILMLN GTRGAVVAML VTSVPMLMIR YRRFSVRMLI VAVCAVATLA IGAYMVPDSP VS KRVDDAV SEIQMYRQGN IETSVGVRLK IWHIGLQYFS EHPWTGVGVG QFARILHASE FCHETKSLAC VLEHAHNDIV EAA STTGIP GLMVMLGLFL VPAVLFARAL RAARSLGNPQ GVSLGGAGLG VVMASLISGL TQVTMAHQAN VVFYAGLIGL LLGM AGREA HSARTDAV UniProtKB: Cell surface polysaccharide polymerase/ligase |
-Macromolecule #2: Fab Heavy (H) Chain
| Macromolecule | Name: Fab Heavy (H) Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.077119 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VAYISSYYGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARIMFKWVSP NMAFDYWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VAYISSYYGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARIMFKWVSP NMAFDYWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHTC |
-Macromolecule #3: Fab Light (L) Chain
| Macromolecule | Name: Fab Light (L) Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.396951 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: GERANYL DIPHOSPHATE
| Macromolecule | Name: GERANYL DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: GPP |
|---|---|
| Molecular weight | Theoretical: 314.209 Da |
| Chemical component information | 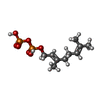 ChemComp-GPP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: PLASMA CLEANING | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 2378 / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.23 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.8) / Number images used: 39844 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)




















