[English] 日本語
 Yorodumi
Yorodumi- EMDB-25927: S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | ||||||||||||
 Map data Map data | unsharpened cryo-EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | initiator / helicase loader / AAA+ ATPase / REPLICATION-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMCM complex loading / CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / Assembly of the ORC complex at the origin of replication / nuclear origin of replication recognition complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / Activation of the pre-replicative complex / nuclear pre-replicative complex / cyclin-dependent protein serine/threonine kinase inhibitor activity ...MCM complex loading / CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / Assembly of the ORC complex at the origin of replication / nuclear origin of replication recognition complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / Activation of the pre-replicative complex / nuclear pre-replicative complex / cyclin-dependent protein serine/threonine kinase inhibitor activity / DNA replication preinitiation complex / Activation of ATR in response to replication stress / nucleosome organization / mitotic DNA replication checkpoint signaling / silent mating-type cassette heterochromatin formation / regulation of DNA-templated DNA replication initiation / CDK-mediated phosphorylation and removal of Cdc6 / Orc1 removal from chromatin / regulation of DNA replication / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / nucleosome binding / G1/S transition of mitotic cell cycle / chromosome / chromosome, telomeric region / cell division / GTPase activity / chromatin binding / GTP binding / ATP hydrolysis activity / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
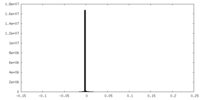
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Schmidt JM / Yang R | ||||||||||||
| Funding support |  United States, European Union, 3 items United States, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6. Authors: Jan Marten Schmidt / Ran Yang / Ashish Kumar / Olivia Hunker / Jan Seebacher / Franziska Bleichert /   Abstract: The coordinated action of multiple replicative helicase loading factors is needed for the licensing of replication origins prior to DNA replication. Binding of the Origin Recognition Complex (ORC) to ...The coordinated action of multiple replicative helicase loading factors is needed for the licensing of replication origins prior to DNA replication. Binding of the Origin Recognition Complex (ORC) to DNA initiates the ATP-dependent recruitment of Cdc6, Cdt1 and Mcm2-7 loading, but the structural details for timely ATPase site regulation and for how loading can be impeded by inhibitory signals, such as cyclin-dependent kinase phosphorylation, are unknown. Using cryo-electron microscopy, we have determined several structures of S. cerevisiae ORC·DNA·Cdc6 intermediates at 2.5-2.7 Å resolution. These structures reveal distinct ring conformations of the initiator·co-loader assembly and inactive ATPase site configurations for ORC and Cdc6. The Orc6 N-terminal domain laterally engages the ORC·Cdc6 ring in a manner that is incompatible with productive Mcm2-7 docking, while deletion of this Orc6 region alleviates the CDK-mediated inhibition of Mcm7 recruitment. Our findings support a model in which Orc6 promotes the assembly of an autoinhibited ORC·DNA·Cdc6 intermediate to block origin licensing in response to CDK phosphorylation and to avert DNA re-replication. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25927.map.gz emd_25927.map.gz | 80.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25927-v30.xml emd-25927-v30.xml emd-25927.xml emd-25927.xml | 35.6 KB 35.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25927_fsc.xml emd_25927_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25927.png emd_25927.png | 111.3 KB | ||
| Filedesc metadata |  emd-25927.cif.gz emd-25927.cif.gz | 9.7 KB | ||
| Others |  emd_25927_additional_1.map.gz emd_25927_additional_1.map.gz emd_25927_additional_2.map.gz emd_25927_additional_2.map.gz emd_25927_half_map_1.map.gz emd_25927_half_map_1.map.gz emd_25927_half_map_2.map.gz emd_25927_half_map_2.map.gz | 10.1 MB 13.9 MB 80.9 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25927 http://ftp.pdbj.org/pub/emdb/structures/EMD-25927 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25927 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25927 | HTTPS FTP |
-Validation report
| Summary document |  emd_25927_validation.pdf.gz emd_25927_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25927_full_validation.pdf.gz emd_25927_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_25927_validation.xml.gz emd_25927_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_25927_validation.cif.gz emd_25927_validation.cif.gz | 23.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25927 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25927 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25927 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25927 | HTTPS FTP |
-Related structure data
| Related structure data |  7tjjMC  7tjfC  7tjhC  7tjiC  7tjkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25927.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25927.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: sharpened, masked cryo-EM map
| File | emd_25927_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened, masked cryo-EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
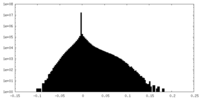
| Density Histograms |
-Additional map: density modified cryo-EM map
| File | emd_25927_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified cryo-EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_25927_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_25927_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : ORC in complex with ARS1 DNA and Cdc6
+Supramolecule #1: ORC in complex with ARS1 DNA and Cdc6
+Macromolecule #1: Origin recognition complex subunit 1
+Macromolecule #2: Origin recognition complex subunit 2
+Macromolecule #3: Origin recognition complex subunit 3
+Macromolecule #4: Origin recognition complex subunit 4
+Macromolecule #5: Origin recognition complex subunit 5
+Macromolecule #6: Origin recognition complex subunit 6
+Macromolecule #9: Cell division control protein 6
+Macromolecule #7: DNA, 84 bp ARS1
+Macromolecule #8: DNA, 84 bp ARS1
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #11: MAGNESIUM ION
+Macromolecule #12: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 6.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)