+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BceAB E169Q variant ATP-bound conformation | |||||||||
 Map data Map data | Cryo-EM map of BceAB in an ATP-bound conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BceAB / bacitracin / transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmembrane transporter activity / transmembrane transport / response to antibiotic / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
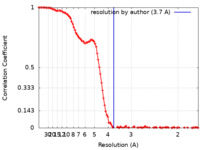
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | George NL / Orlando BJ | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Conformational snapshots of the bacitracin sensing and resistance transporter BceAB. Authors: Natasha L George / Anthony L Schilmiller / Benjamin J Orlando /  Abstract: SignificanceMany gram-positive organisms have evolved an elegant solution to sense and resist antimicrobial peptides that inhibit cell-wall synthesis. These organisms express an unusual "Bce-type" ...SignificanceMany gram-positive organisms have evolved an elegant solution to sense and resist antimicrobial peptides that inhibit cell-wall synthesis. These organisms express an unusual "Bce-type" adenosine triphosphate-binding cassette (ABC) transporter that recognizes complexes formed between antimicrobial peptides and lipids involved in cell-wall biosynthesis. In this work, we provide the first structural snapshots of a Bce-type ABC transporter trapped in different conformational states. Our structures and associated biochemical data provide key insights into the novel target protection mechanism that these unusual ABC transporters use to sense and resist antimicrobial peptides. The studies described herein set the stage to begin developing a comprehensive molecular understanding of the diverse interactions between antimicrobial peptides and conserved resistance machinery found across most gram-positive organisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25812.map.gz emd_25812.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25812-v30.xml emd-25812-v30.xml emd-25812.xml emd-25812.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25812_fsc.xml emd_25812_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_25812.png emd_25812.png | 29.9 KB | ||
| Filedesc metadata |  emd-25812.cif.gz emd-25812.cif.gz | 6.4 KB | ||
| Others |  emd_25812_half_map_1.map.gz emd_25812_half_map_1.map.gz emd_25812_half_map_2.map.gz emd_25812_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25812 http://ftp.pdbj.org/pub/emdb/structures/EMD-25812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25812 | HTTPS FTP |
-Related structure data
| Related structure data |  7tchMC  7tcgC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25812.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25812.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of BceAB in an ATP-bound conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.872 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map A
| File | emd_25812_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_25812_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BceAB (E169Q variant)
| Entire | Name: BceAB (E169Q variant) |
|---|---|
| Components |
|
-Supramolecule #1: BceAB (E169Q variant)
| Supramolecule | Name: BceAB (E169Q variant) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 129 KDa |
-Macromolecule #1: Bacitracin export permease protein BceB
| Macromolecule | Name: Bacitracin export permease protein BceB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: 168 |
| Molecular weight | Theoretical: 72.262109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNINQLILRN LKKNLRNYYL YVFALIFSVA LYFAFVTLQY DPAINEVKAS IKGAAAIKTA SILLVAVVAI FILYANTIFI KRRSKEIGL FQLIGMTKHK IFRILSAENV MLYFGSLAIG VAAGFSISKL VLMILFKIVD VKADAKLHFS EQALVQTVIV F CGIYLLIM ...String: MNINQLILRN LKKNLRNYYL YVFALIFSVA LYFAFVTLQY DPAINEVKAS IKGAAAIKTA SILLVAVVAI FILYANTIFI KRRSKEIGL FQLIGMTKHK IFRILSAENV MLYFGSLAIG VAAGFSISKL VLMILFKIVD VKADAKLHFS EQALVQTVIV F CGIYLLIM IMNYTFIKKQ SILSLFKVTS STEDKVKKIS FFQMLIGALG IVLILTGYYV SSELFGGKFK TINELFVAMS FI LGSVIIG TFLFYKGSVT FISNIIRKSK GGYLNISEVL SLSSIMFRMK SNALLLTIIT TVSALAIGLL SLAYISYYSS EKT AEQNVA ADFSFMNEKD AKLFENKLRE SNISFVKKAT PVLQANVDIA NIMDGTPKEM QGDPGNMQLA VVSDKDVKGV DVAA GEAVF SGYTDLLQKI MVFKDSGVIK VKSKHETQPL KYKGLREEFL VSYTFTSGGM PAVIVDDSLF KQLDKDKDPR IQLAQ STFI GVNVKHDDQM EKANELFQQV NKKNEHLSRL DTSAAQKSLF GMVMFIVGFL GLTFLITSGC ILYFKQMGES EDEKPS YTI LRKLGFTQGD LIKGIRIKQM YNFGIPLVVG LFHSYFAVQS GWFLFGSEVW APMIMVMVLY TALYSIFGFL SVLYYKK VI KSSL UniProtKB: Bacitracin export permease protein BceB |
-Macromolecule #2: Bacitracin export ATP-binding protein BceA
| Macromolecule | Name: Bacitracin export ATP-binding protein BceA / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: 168 |
| Molecular weight | Theoretical: 29.247393 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGHHHHHHV ILEANKIRKS YGNKLNKQEV LKGIDIHIEK GEFVSIMGAS GSGKTTLLNV LSSIDQVSHG TIHINGNDMT AMKEKQLAE FRKQHLGFIF QDYNLLDTLT VKENILLPLS ITKLSKKEAN RKFEEVAKEL GIYELRDKYP NEISGGQKQR T SAGRAFIH ...String: MSGHHHHHHV ILEANKIRKS YGNKLNKQEV LKGIDIHIEK GEFVSIMGAS GSGKTTLLNV LSSIDQVSHG TIHINGNDMT AMKEKQLAE FRKQHLGFIF QDYNLLDTLT VKENILLPLS ITKLSKKEAN RKFEEVAKEL GIYELRDKYP NEISGGQKQR T SAGRAFIH DPSIIFADQP TGALDSKSAS DLLNKLSQLN QKRNATIIMV THDPVAASYC GRVIFIKDGQ MYTQLNKGGQ DR QTFFQDI MKTQGVLGGV QHEH UniProtKB: Bacitracin export ATP-binding protein BceA |
-Macromolecule #3: 4-amino-4-deoxy-1-O-[(S)-hydroxy{[(2E,6E,10Z,14Z,18Z,22E,26E,30E)...
| Macromolecule | Name: 4-amino-4-deoxy-1-O-[(S)-hydroxy{[(2E,6E,10Z,14Z,18Z,22E,26E,30E)-3,7,11,15,19,23,27,31,35-nonamethylhexatriaconta-2,6,10,14,18,22,26,30,34-nonaen-1-yl]oxy}phosphoryl]-alpha-L-arabinopyranose type: ligand / ID: 3 / Number of copies: 1 / Formula: I0O |
|---|---|
| Molecular weight | Theoretical: 842.178 Da |
| Chemical component information |  ChemComp-I0O: |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 15mA in Pelco EasyGlow | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.1 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7tch: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)