[English] 日本語
 Yorodumi
Yorodumi- EMDB-25795: Structure of In-vitro Synthesized Cellulose Fibrils by Averaging ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of In-vitro Synthesized Cellulose Fibrils by Averaging non-overlapping subtograms of 10.9x26.5x10.9 nm | |||||||||
 Map data Map data | Average of 10.9x26.5x10.9 nm subtograms of in-vitro synthesized cellulose fibrils. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cellulose / microfibril / in-vitro synthesized / CARBOHYDRATE | |||||||||
| Biological species |  Physcomitrium patens (plant) Physcomitrium patens (plant) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Nixon BT / Frank MA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biomacromolecules / Year: 2022 Journal: Biomacromolecules / Year: 2022Title: Structure of -Synthesized Cellulose Fibrils Viewed by Cryo-Electron Tomography and C Natural-Abundance Dynamic Nuclear Polarization Solid-State NMR. Authors: Fabien Deligey / Mark A Frank / Sung Hyun Cho / Alex Kirui / Frederic Mentink-Vigier / Matthew T Swulius / B Tracy Nixon / Tuo Wang /  Abstract: Cellulose, the most abundant biopolymer, is a central source for renewable energy and functionalized materials. synthesis of cellulose microfibrils (CMFs) has become possible using purified ...Cellulose, the most abundant biopolymer, is a central source for renewable energy and functionalized materials. synthesis of cellulose microfibrils (CMFs) has become possible using purified cellulose synthase (CESA) isoforms from and hybrid aspen. The exact nature of these fibrils remains unknown. Here, we characterize -synthesized fibers made by CESAs present in membrane fractions of over-expressing CESA5 by cryo-electron tomography and dynamic nuclear polarization (DNP) solid-state NMR. DNP enabled measuring two-dimensional C-C correlation spectra without isotope-labeling of the fibers. Results show structural similarity between fibrils and native CMF in plant cell walls. Intensity quantifications agree with the 18-chain structural model for plant CMF and indicate limited fibrillar bundling. The system thus reveals insights into cell wall synthesis and may contribute to novel cellulosic materials. The integrated DNP and cryo-electron tomography methods are also applicable to structural studies of other carbohydrate-based biomaterials. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25795.map.gz emd_25795.map.gz | 904.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25795-v30.xml emd-25795-v30.xml emd-25795.xml emd-25795.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25795.png emd_25795.png | 48.4 KB | ||
| Filedesc metadata |  emd-25795.cif.gz emd-25795.cif.gz | 4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25795 http://ftp.pdbj.org/pub/emdb/structures/EMD-25795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25795 | HTTPS FTP |
-Validation report
| Summary document |  emd_25795_validation.pdf.gz emd_25795_validation.pdf.gz | 384.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25795_full_validation.pdf.gz emd_25795_full_validation.pdf.gz | 384 KB | Display | |
| Data in XML |  emd_25795_validation.xml.gz emd_25795_validation.xml.gz | 4.7 KB | Display | |
| Data in CIF |  emd_25795_validation.cif.gz emd_25795_validation.cif.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25795 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25795 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25795 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25795 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25795.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25795.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Average of 10.9x26.5x10.9 nm subtograms of in-vitro synthesized cellulose fibrils. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.102 Å | ||||||||||||||||||||||||||||||||||||
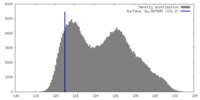
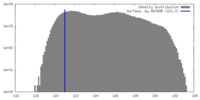
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : In-vitro synthesized cellulose microfibril
| Entire | Name: In-vitro synthesized cellulose microfibril |
|---|---|
| Components |
|
-Supramolecule #1: In-vitro synthesized cellulose microfibril
| Supramolecule | Name: In-vitro synthesized cellulose microfibril / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Physcomitrium patens (plant) Physcomitrium patens (plant) |
| Molecular weight | Theoretical: 5.4 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 3.658 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -5.0 µm / Nominal defocus min: -5.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET (ver. 1.15.1) / Number subtomograms used: 4374 |
|---|---|
| Extraction | Number tomograms: 12 / Number images used: 4384 |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name:  IMOD (ver. 4.12.8) / Software - details: 3dmod IMOD (ver. 4.12.8) / Software - details: 3dmod |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)