+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
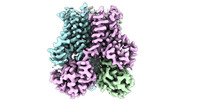
| Title | Structure of STEAP2 in complex with ligands | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane-embedded oxidoreductase / membrane protein / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcopper ion import across plasma membrane / Oxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor / ferric-chelate reductase (NADPH) activity / iron ion import across plasma membrane / cupric reductase (NADH) activity / copper ion import / regulated exocytosis / trans-Golgi network transport vesicle / Golgi to plasma membrane transport / response to hormone ...copper ion import across plasma membrane / Oxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor / ferric-chelate reductase (NADPH) activity / iron ion import across plasma membrane / cupric reductase (NADH) activity / copper ion import / regulated exocytosis / trans-Golgi network transport vesicle / Golgi to plasma membrane transport / response to hormone / endocytosis / early endosome / endosome / endosome membrane / Golgi membrane / metal ion binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wang L / Chen KH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Mechanism of stepwise electron transfer in six-transmembrane epithelial antigen of the prostate (STEAP) 1 and 2. Authors: Kehan Chen / Lie Wang / Jiemin Shen / Ah-Lim Tsai / Ming Zhou / Gang Wu /  Abstract: Six transmembrane epithelial antigen of the prostate (STEAP) 1-4 are membrane-embedded hemoproteins that chelate a heme prosthetic group in a transmembrane domain (TMD). STEAP2-4, but not STEAP1, ...Six transmembrane epithelial antigen of the prostate (STEAP) 1-4 are membrane-embedded hemoproteins that chelate a heme prosthetic group in a transmembrane domain (TMD). STEAP2-4, but not STEAP1, have an intracellular oxidoreductase domain (OxRD) and can mediate cross-membrane electron transfer from NADPH via FAD and heme. However, it is unknown whether STEAP1 can establish a physiologically relevant electron transfer chain. Here, we show that STEAP1 can be reduced by reduced FAD or soluble cytochrome reductase that serves as a surrogate OxRD, providing the first evidence that STEAP1 can support a cross-membrane electron transfer chain. It is not clear whether FAD, which relays electrons from NADPH in OxRD to heme in TMD, remains constantly bound to the STEAPs. We found that FAD reduced by STEAP2 can be utilized by STEAP1, suggesting that FAD is diffusible rather than staying bound to STEAP2. We determined the structure of human STEAP2 in complex with NADP and FAD to an overall resolution of 3.2 Å by cryo-electron microscopy and found that the two cofactors bind STEAP2 similarly as in STEAP4, suggesting that a diffusible FAD is a general feature of the electron transfer mechanism in the STEAPs. We also demonstrated that STEAP2 reduces ferric nitrilotriacetic acid (Fe-NTA) significantly slower than STEAP1 and proposed that the slower reduction is due to the poor Fe-NTA binding to the highly flexible extracellular region in STEAP2. These results establish a solid foundation for understanding the function and mechanisms of the STEAPs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25775.map.gz emd_25775.map.gz | 26.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25775-v30.xml emd-25775-v30.xml emd-25775.xml emd-25775.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25775.png emd_25775.png | 66 KB | ||
| Filedesc metadata |  emd-25775.cif.gz emd-25775.cif.gz | 6.6 KB | ||
| Others |  emd_25775_half_map_1.map.gz emd_25775_half_map_1.map.gz emd_25775_half_map_2.map.gz emd_25775_half_map_2.map.gz | 48.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25775 http://ftp.pdbj.org/pub/emdb/structures/EMD-25775 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25775 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25775 | HTTPS FTP |
-Related structure data
| Related structure data |  7taiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25775.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25775.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_25775_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25775_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homo-trimeric complex of STEAP2
| Entire | Name: Homo-trimeric complex of STEAP2 |
|---|---|
| Components |
|
-Supramolecule #1: Homo-trimeric complex of STEAP2
| Supramolecule | Name: Homo-trimeric complex of STEAP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56 KDa |
-Macromolecule #1: Metalloreductase STEAP2
| Macromolecule | Name: Metalloreductase STEAP2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO EC number: Oxidoreductases; Oxidizing metal ions; With NAD+ or NADP+ as acceptor |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.115305 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MESISMMGSP KSLSETFLPN GINGIKDARK VTVGVIGSGD FAKSLTIRLI RCGYHVVIGS RNPKFASEFF PHVVDVTHHE DALTKTNII FVAIHREHYT SLWDLRHLLV GKILIDVSNN MRINQYPESN AEYLASLFPD SLIVKGFNVV SAWALQLGPK D ASRQVYIC ...String: MESISMMGSP KSLSETFLPN GINGIKDARK VTVGVIGSGD FAKSLTIRLI RCGYHVVIGS RNPKFASEFF PHVVDVTHHE DALTKTNII FVAIHREHYT SLWDLRHLLV GKILIDVSNN MRINQYPESN AEYLASLFPD SLIVKGFNVV SAWALQLGPK D ASRQVYIC SNNIQARQQV IELARQLNFI PIDLGSLSSA REIENLPLRL FTLWRGPVVV AISLATFFFL YSFVRDVIHP YA RNQQSDF YKIPIEIVNK TLPIVAITLL SLVYLAGLLA AAYQLYYGTK YRRFPPWLET WLQCRKQLGL LSFFFAMVHV AYS LCLPMR RSERYLFLNM AYQQVHANIE NSWNEEEVWR IEMYISFGIM SLGLLSLLAV TSIPSVSNAL NWREFSFIQS TLGY VALLI STFHVLIYGW KRAFEEEYYR FYTPPNFVLA LVLPSIVILG KIILFLPCIS RKLKRIKKGW EKSQFLEEGM GGTIP HVSP ERVTVM UniProtKB: Metalloreductase STEAP2 |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 3 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 3 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #4: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAP |
|---|---|
| Molecular weight | Theoretical: 743.405 Da |
| Chemical component information |  ChemComp-NAP: |
-Macromolecule #5: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 5 / Number of copies: 3 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 6 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































