[English] 日本語
 Yorodumi
Yorodumi- EMDB-25773: Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | |||||||||
 Map data Map data | Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virus / EV-D68 / acute flaccid myelitis / AFM / inhibitor / antiviral / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID / VIRUS-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpicornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport ...picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / RNA helicase activity / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  enterovirus D68 enterovirus D68 | |||||||||
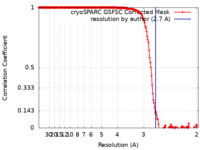
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Fu J / Klose T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Isoxazole-3-Carboxamide Derivatives of Pleconaril Destabilize the Viral Capsid of Enterovirus-D68 Authors: Lane T / Fu J / Sherry B / Tarbet B / Hurst BL / Riabova O / Kazakova E / Egorova A / Clarke P / Leser S / Frost J / Rudy M / Tyler K / Klose T / Kuhn RJ / Makarov V / Ekins S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25773.map.gz emd_25773.map.gz | 483.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25773-v30.xml emd-25773-v30.xml emd-25773.xml emd-25773.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25773_fsc.xml emd_25773_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_25773.png emd_25773.png | 297.7 KB | ||
| Filedesc metadata |  emd-25773.cif.gz emd-25773.cif.gz | 6.5 KB | ||
| Others |  emd_25773_half_map_1.map.gz emd_25773_half_map_1.map.gz emd_25773_half_map_2.map.gz emd_25773_half_map_2.map.gz | 472.4 MB 472.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25773 http://ftp.pdbj.org/pub/emdb/structures/EMD-25773 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25773 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25773 | HTTPS FTP |
-Validation report
| Summary document |  emd_25773_validation.pdf.gz emd_25773_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25773_full_validation.pdf.gz emd_25773_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_25773_validation.xml.gz emd_25773_validation.xml.gz | 25 KB | Display | |
| Data in CIF |  emd_25773_validation.cif.gz emd_25773_validation.cif.gz | 32.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25773 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25773 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25773 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25773 | HTTPS FTP |
-Related structure data
| Related structure data |  7tagMC  7t9pC  7tafC  7tahC  7tajC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25773.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25773.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.98044 Å | ||||||||||||||||||||||||||||||||||||
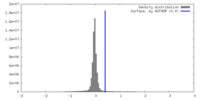
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Enterovirus D68 US/MO/14-18947 strain virion in complex with...
| File | emd_25773_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | ||||||||||||
| Projections & Slices |
| ||||||||||||
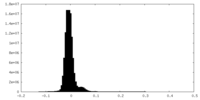
| Density Histograms |
-Half map: Enterovirus D68 US/MO/14-18947 strain virion in complex with...
| File | emd_25773_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | ||||||||||||
| Projections & Slices |
| ||||||||||||
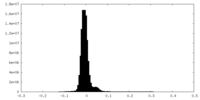
| Density Histograms |
- Sample components
Sample components
-Entire : enterovirus D68
| Entire | Name:  enterovirus D68 enterovirus D68 |
|---|---|
| Components |
|
-Supramolecule #1: enterovirus D68
| Supramolecule | Name: enterovirus D68 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 42789 / Sci species name: enterovirus D68 / Sci species strain: US/MO/14-18947 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: viral protein 1
| Macromolecule | Name: viral protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  enterovirus D68 / Strain: US/MO/14-18947 enterovirus D68 / Strain: US/MO/14-18947 |
| Molecular weight | Theoretical: 32.819207 KDa |
| Sequence | String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSTARADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ...String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSTARADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ASVFFKISDP PARITIPFMC INSAYSVFYD GFAGFEKNGL YGINPADTIG NLCVRIVNEH QPVGFTVTVR VY MKPKHIK AWAPRPPRTL PYMSIANANY KGKERAPNAL SAIIGNRDSV KTMPHNIVN UniProtKB: Genome polyprotein |
-Macromolecule #2: viral protein 3
| Macromolecule | Name: viral protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  enterovirus D68 / Strain: US/MO/14-18947 enterovirus D68 / Strain: US/MO/14-18947 |
| Molecular weight | Theoretical: 27.112814 KDa |
| Sequence | String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMAAGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP ...String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMAAGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP WISGSHYRMF NNDAKSTNAN VGYVTCFMQT NLIVPSESSD TCSLIGFIAA KDDFSLRLMR DSPDIGQLDH LH AAEAAYQ UniProtKB: Genome polyprotein |
-Macromolecule #3: viral protein 2
| Macromolecule | Name: viral protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  enterovirus D68 / Strain: US/MO/14-18947 enterovirus D68 / Strain: US/MO/14-18947 |
| Molecular weight | Theoretical: 26.573094 KDa |
| Sequence | String: SDRVLQLKLG NSAIVTQEAA NYCCAYGEWP NYLPDHEAVA IDKPTQPETA TDRFYTLKSV KWETGSTGWW WKLPDALNNI GMFGQNVQH HYLYRSGFLI HVQCNATKFH QGALLVVAIP EHQRGAHNTN TSPGFDDIMK GEEGGTFNHP YVLDDGTSLA C ATIFPHQW ...String: SDRVLQLKLG NSAIVTQEAA NYCCAYGEWP NYLPDHEAVA IDKPTQPETA TDRFYTLKSV KWETGSTGWW WKLPDALNNI GMFGQNVQH HYLYRSGFLI HVQCNATKFH QGALLVVAIP EHQRGAHNTN TSPGFDDIMK GEEGGTFNHP YVLDDGTSLA C ATIFPHQW INLRTNNSAT IVLPWMNAAP MDFPLRHNQW TLAIIPVVPL GTRTTSSMVP ITVSIAPMCC EFNGLRHAIT UniProtKB: Genome polyprotein |
-Macromolecule #4: viral protein 4
| Macromolecule | Name: viral protein 4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  enterovirus D68 / Strain: US/MO/14-18947 enterovirus D68 / Strain: US/MO/14-18947 |
| Molecular weight | Theoretical: 7.33696 KDa |
| Sequence | String: GAQVTRQQTG THENANIATN GSHITYNQIN FYKDSYAASA SKQDFSQDPS KFTEPVVEGL KAGAPVLK UniProtKB: Genome polyprotein |
-Macromolecule #5: 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-...
| Macromolecule | Name: 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE type: ligand / ID: 5 / Number of copies: 1 / Formula: W11 |
|---|---|
| Molecular weight | Theoretical: 381.349 Da |
| Chemical component information | 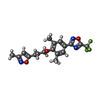 ChemComp-W11: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 36.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


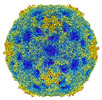






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)