+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
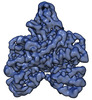
| Title | human DNA polymerase gamma accessory subunit, POLG2 | ||||||||||||
 Map data Map data | human DNA polymerase gamma accessory subunit, POLG2 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | processivity factor / accessory subunit / DNA BINDING PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Riccio AA / Bouvette J / Borgnia MJ / Copeland WC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Automated systematic evaluation of cryo-EM specimens with SmartScope. Authors: Jonathan Bouvette / Qinwen Huang / Amanda A Riccio / William C Copeland / Alberto Bartesaghi / Mario J Borgnia /  Abstract: Finding the conditions to stabilize a macromolecular target for imaging remains the most critical barrier to determining its structure by cryo-electron microscopy (cryo-EM). While automation has ...Finding the conditions to stabilize a macromolecular target for imaging remains the most critical barrier to determining its structure by cryo-electron microscopy (cryo-EM). While automation has significantly increased the speed of data collection, specimens are still screened manually, a laborious and subjective task that often determines the success of a project. Here, we present SmartScope, the first framework to streamline, standardize, and automate specimen evaluation in cryo-EM. SmartScope employs deep-learning-based object detection to identify and classify features suitable for imaging, allowing it to perform thorough specimen screening in a fully automated manner. A web interface provides remote control over the automated operation of the microscope in real time and access to images and annotation tools. Manual annotations can be used to re-train the feature recognition models, leading to improvements in performance. Our automated tool for systematic evaluation of specimens streamlines structure determination and lowers the barrier of adoption for cryo-EM. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25764.map.gz emd_25764.map.gz | 56.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25764-v30.xml emd-25764-v30.xml emd-25764.xml emd-25764.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25764_fsc.xml emd_25764_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25764.png emd_25764.png | 162.2 KB | ||
| Filedesc metadata |  emd-25764.cif.gz emd-25764.cif.gz | 4.9 KB | ||
| Others |  emd_25764_half_map_1.map.gz emd_25764_half_map_1.map.gz emd_25764_half_map_2.map.gz emd_25764_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25764 http://ftp.pdbj.org/pub/emdb/structures/EMD-25764 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25764 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25764 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25764.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25764.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human DNA polymerase gamma accessory subunit, POLG2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 2
| File | emd_25764_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
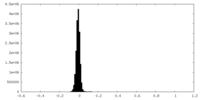
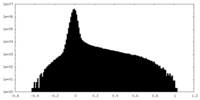
| Density Histograms |
-Half map: Half Map 1
| File | emd_25764_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
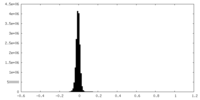
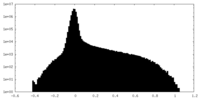
| Density Histograms |
- Sample components
Sample components
-Entire : human DNA polymerase gamma accessory subunit, POLG2
| Entire | Name: human DNA polymerase gamma accessory subunit, POLG2 |
|---|---|
| Components |
|
-Supramolecule #1: human DNA polymerase gamma accessory subunit, POLG2
| Supramolecule | Name: human DNA polymerase gamma accessory subunit, POLG2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54 KDa |
-Macromolecule #1: human DNA polymerase gamma accessory subunit, POLG2
| Macromolecule | Name: human DNA polymerase gamma accessory subunit, POLG2 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRGSHHHHHH GSTMDAGQPE LLTERSSPKG GHVKSHAELE GNGEHPEAPG SGEG SEALL EICQRRHFLS GSKQQLSRDS LLSGCHPGFG PLGVELRKNL AAEWWTSVVV FREQVFPVDA LHHKPGPLLP GDSAFRLVSA ETLREILQDK ELSKEQLVTF LENVLKTSGK ...String: MRGSHHHHHH GSTMDAGQPE LLTERSSPKG GHVKSHAELE GNGEHPEAPG SGEG SEALL EICQRRHFLS GSKQQLSRDS LLSGCHPGFG PLGVELRKNL AAEWWTSVVV FREQVFPVDA LHHKPGPLLP GDSAFRLVSA ETLREILQDK ELSKEQLVTF LENVLKTSGK LRENLLHGAL EHYVNCLDLV NKRLPYGLAQ IGVCFHPVFD TKQIRNGVKS IGEKTEASLV WFTPPRTSNQ WLDFWLRHRL QWWRKFAMSP SNFSSSDCQD EEGRKGNKLY YNFPWGKELI ETLWNLGDHE LLHMYPGNVS KLHGRDGRKN VVPCVLSVNG DLDRGMLAYL YDSFQLTENS FTRKKNLHRK VLKLHPCLAP IKVALDVGRG PTLELRQVCQ GLFNELLENG ISVWPGYLET MQSSLEQLYS KYDEMSILFT VLVTETTLEN GLIHLRSRDT TMKEMMHISK LKDFLIKYIS SAKNV |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.22 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3708 pixel / Digitization - Dimensions - Height: 3838 pixel / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)