+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of VcINDY-apo | ||||||||||||||||||
 Map data Map data | Phenix Sharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | TRANSPORTER / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology | Citrate transporter-like domain / Citrate transporter / Sodium/sulphate symporter, conserved site / Sodium:sulfate symporter family signature. / succinate transmembrane transporter activity / Solute carrier family 13 / plasma membrane / Transporter, NadC family Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
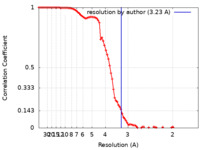
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | ||||||||||||||||||
 Authors Authors | Sauer DB / Marden JJ | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of ion - substrate coupling in the Na-dependent dicarboxylate transporter VcINDY. Authors: David B Sauer / Jennifer J Marden / Joseph C Sudar / Jinmei Song / Christopher Mulligan / Da-Neng Wang /   Abstract: The Na-dependent dicarboxylate transporter from Vibrio cholerae (VcINDY) is a prototype for the divalent anion sodium symporter (DASS) family. While the utilization of an electrochemical Na gradient ...The Na-dependent dicarboxylate transporter from Vibrio cholerae (VcINDY) is a prototype for the divalent anion sodium symporter (DASS) family. While the utilization of an electrochemical Na gradient to power substrate transport is well established for VcINDY, the structural basis of this coupling between sodium and substrate binding is not currently understood. Here, using a combination of cryo-EM structure determination, succinate binding and site-directed cysteine alkylation assays, we demonstrate that the VcINDY protein couples sodium- and substrate-binding via a previously unseen cooperative mechanism by conformational selection. In the absence of sodium, substrate binding is abolished, with the succinate binding regions exhibiting increased flexibility, including HPb, TM10b and the substrate clamshell motifs. Upon sodium binding, these regions become structurally ordered and create a proper binding site for the substrate. Taken together, these results provide strong evidence that VcINDY's conformational selection mechanism is a result of the sodium-dependent formation of the substrate binding site. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25756.map.gz emd_25756.map.gz | 58.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25756-v30.xml emd-25756-v30.xml emd-25756.xml emd-25756.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25756_fsc.xml emd_25756_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_25756.png emd_25756.png | 80.7 KB | ||
| Filedesc metadata |  emd-25756.cif.gz emd-25756.cif.gz | 5.8 KB | ||
| Others |  emd_25756_additional_1.map.gz emd_25756_additional_1.map.gz emd_25756_half_map_1.map.gz emd_25756_half_map_1.map.gz emd_25756_half_map_2.map.gz emd_25756_half_map_2.map.gz | 31.9 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25756 http://ftp.pdbj.org/pub/emdb/structures/EMD-25756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25756 | HTTPS FTP |
-Related structure data
| Related structure data |  7t9fMC  7t9gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10970 (Title: Particle stacks for VcINDY in Choline Chloride / Data size: 383.6 EMPIAR-10970 (Title: Particle stacks for VcINDY in Choline Chloride / Data size: 383.6 Data #1: Particle stack of VcINDY in Choline Chloride Class 1 [picked particles - multiframe - processed] Data #2: Particle stack of VcINDY in Choline Chloride Class 1 [picked particles - multiframe - processed] Data #3: Particle stack of VcINDY in Choline Chloride Class 3 [picked particles - multiframe - processed] Data #4: Particle stack of VcINDY in Choline Chloride Class 4 [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25756.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25756.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_25756_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap A
| File | emd_25756_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap B
| File | emd_25756_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VcINDY in the Ci-apo state
| Entire | Name: VcINDY in the Ci-apo state |
|---|---|
| Components |
|
-Supramolecule #1: VcINDY in the Ci-apo state
| Supramolecule | Name: VcINDY in the Ci-apo state / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DASS family sodium-coupled anion symporter
| Macromolecule | Name: DASS family sodium-coupled anion symporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.157359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: REWFLHRNSL IVLADVALFL ALYHFLPFEH NVVLGISMLA FIAVLWLTEA LHVTVTAILV PVMAVFFGIF ETQAALNNFA NSIIFLFLG GFALAAAMHH QGLDKVIADK VLAMAQGKMS VAVFMLFGVT ALLSMWISNT ATAAMMLPLV LGVLSKVDAD K QRSTYVFV ...String: REWFLHRNSL IVLADVALFL ALYHFLPFEH NVVLGISMLA FIAVLWLTEA LHVTVTAILV PVMAVFFGIF ETQAALNNFA NSIIFLFLG GFALAAAMHH QGLDKVIADK VLAMAQGKMS VAVFMLFGVT ALLSMWISNT ATAAMMLPLV LGVLSKVDAD K QRSTYVFV LLGVAYSASI GGIATLVGSP PNAIAAAEVG LSFTDWMKFG LPTAMMMLPM AIAILYFLLK PTLNGMFELD RA PVNWDKG KVVTLGIFGL TVFLWIFSSP INAALGGFKS FDTLVALGAI LMLSFARVVH WKEIQKTADW GVLLLFGGGL CLS NVLKQT GTSVFLANAL SDMVSHMGIF VVILVVATFV VFLTEFASNT ASAALLIPVF ATVAEAFGMS PVLLSVLIAV AASC AFMLP VATPPNAIVF ASGHIKQSEM MRVGLYLNIA CIGLLTAIAM LFWQ UniProtKB: Transporter, NadC family |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4514 / Average exposure time: 2.34 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)