+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of NBD-ffsy filaments (class 1) | |||||||||||||||
 Map data Map data | Cryo-EM of NBD-ffsy filaments (class 1) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | transcytosis / hydrogel / peptides / nanofibers / spheroids / self-assembly peptide filament / PROTEIN FIBRIL | |||||||||||||||
| Biological species | Synthetic construct (others) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Wang F / Guo J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2023 Journal: Nat Nanotechnol / Year: 2023Title: Cell spheroid creation by transcytotic intercellular gelation. Authors: Jiaqi Guo / Fengbin Wang / Yimeng Huang / Hongjian He / Weiyi Tan / Meihui Yi / Edward H Egelman / Bing Xu /  Abstract: Cell spheroids bridge the discontinuity between in vitro systems and in vivo animal models. However, inducing cell spheroids by nanomaterials remains an inefficient and poorly understood process. ...Cell spheroids bridge the discontinuity between in vitro systems and in vivo animal models. However, inducing cell spheroids by nanomaterials remains an inefficient and poorly understood process. Here we use cryogenic electron microscopy to determine the atomic structure of helical nanofibres self-assembled from enzyme-responsive D-peptides and fluorescent imaging to show that the transcytosis of D-peptides induces intercellular nanofibres/gels that potentially interact with fibronectin to enable cell spheroid formation. Specifically, D-phosphopeptides, being protease resistant, undergo endocytosis and endosomal dephosphorylation to generate helical nanofibres. On secretion to the cell surface, these nanofibres form intercellular gels that act as artificial matrices and facilitate the fibrillogenesis of fibronectins to induce cell spheroids. No spheroid formation occurs without endo- or exocytosis, phosphate triggers or shape switching of the peptide assemblies. This study-coupling transcytosis and morphological transformation of peptide assemblies-demonstrates a potential approach for regenerative medicine and tissue engineering. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25714.map.gz emd_25714.map.gz | 65 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25714-v30.xml emd-25714-v30.xml emd-25714.xml emd-25714.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25714.png emd_25714.png | 139.8 KB | ||
| Filedesc metadata |  emd-25714.cif.gz emd-25714.cif.gz | 4.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25714 http://ftp.pdbj.org/pub/emdb/structures/EMD-25714 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25714 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25714 | HTTPS FTP |
-Validation report
| Summary document |  emd_25714_validation.pdf.gz emd_25714_validation.pdf.gz | 526.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25714_full_validation.pdf.gz emd_25714_full_validation.pdf.gz | 526.5 KB | Display | |
| Data in XML |  emd_25714_validation.xml.gz emd_25714_validation.xml.gz | 6.8 KB | Display | |
| Data in CIF |  emd_25714_validation.cif.gz emd_25714_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25714 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25714 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25714 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25714 | HTTPS FTP |
-Related structure data
| Related structure data |  7t6eMC  8dstC  8fofC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25714.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25714.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM of NBD-ffsy filaments (class 1) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
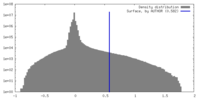
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : self-assemble NBD-ffsy nanotube
| Entire | Name: self-assemble NBD-ffsy nanotube |
|---|---|
| Components |
|
-Supramolecule #1: self-assemble NBD-ffsy nanotube
| Supramolecule | Name: self-assemble NBD-ffsy nanotube / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) / Synthetically produced: Yes |
-Macromolecule #1: NBD-ffsy peptide
| Macromolecule | Name: NBD-ffsy peptide / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Molecular weight | Theoretical: 796.783 Da |
| Sequence | String: (UQ4)(DPN)(DPN)(DSN)(DTY) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 0.483 Å Applied symmetry - Helical parameters - Δ&Phi: 35.77 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: OTHER / Details: map:map FSC and model:map FSC / Number images used: 677537 |
|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)