+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the YejM/LapB complex | |||||||||
 Map data Map data | sharpened map with a B factor of -216 A2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | YejM / YciM / regulation / lipopolysaccharide synthesis / LpxC / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlipopolysaccharide metabolic process / regulation of lipid biosynthetic process / cytoplasmic side of plasma membrane / iron ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
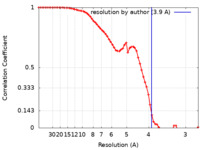
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Mi W / Shu S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli. Authors: Sheng Shu / Wei Mi /  Abstract: Lipopolysaccharide (LPS) is an essential glycolipid and forms a protective permeability barrier for most Gram-negative bacteria. In E. coli, LPS levels are under feedback control, achieved by FtsH- ...Lipopolysaccharide (LPS) is an essential glycolipid and forms a protective permeability barrier for most Gram-negative bacteria. In E. coli, LPS levels are under feedback control, achieved by FtsH-mediated degradation of LpxC, which catalyzes the first committed step in LPS synthesis. FtsH is a membrane-bound AAA+ protease, and its protease activity toward LpxC is regulated by essential membrane proteins LapB and YejM. However, the regulatory mechanisms are elusive. We establish an in vitro assay to analyze the kinetics of LpxC degradation and demonstrate that LapB is an adaptor protein that utilizes its transmembrane helix to interact with FtsH and its cytoplasmic domains to recruit LpxC. Our YejM/LapB complex structure reveals that YejM is an anti-adaptor protein, competing with FtsH for LapB to inhibit LpxC degradation. Structural analysis unravels that LapB and LPS have overlapping binding sites in YejM. Thus, LPS levels control formation of the YejM/LapB complex to determine LpxC protein levels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25713.map.gz emd_25713.map.gz | 25.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25713-v30.xml emd-25713-v30.xml emd-25713.xml emd-25713.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25713_fsc.xml emd_25713_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25713.png emd_25713.png | 77.8 KB | ||
| Masks |  emd_25713_msk_1.map emd_25713_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25713.cif.gz emd-25713.cif.gz | 11.1 KB | ||
| Others |  emd_25713_additional_1.map.gz emd_25713_additional_1.map.gz emd_25713_half_map_1.map.gz emd_25713_half_map_1.map.gz emd_25713_half_map_2.map.gz emd_25713_half_map_2.map.gz | 13.4 MB 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25713 http://ftp.pdbj.org/pub/emdb/structures/EMD-25713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25713 | HTTPS FTP |
-Related structure data
| Related structure data |  7t6dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25713.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25713.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map with a B factor of -216 A2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.346 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25713_msk_1.map emd_25713_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: original 3D reconstruction without sharpening
| File | emd_25713_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | original 3D reconstruction without sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25713_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25713_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : complex of YejM and YciM
| Entire | Name: complex of YejM and YciM |
|---|---|
| Components |
|
-Supramolecule #1: complex of YejM and YciM
| Supramolecule | Name: complex of YejM and YciM / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 225.556 kDa/nm |
-Macromolecule #1: Lipopolysaccharide assembly protein B
| Macromolecule | Name: Lipopolysaccharide assembly protein B / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.546953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLELLFLLLP VAAAYGWYMG RRSAQQNKQD EANRLSRDYV AGVNFLLSNQ QDKAVDLFLD MLKEDTGTVE AHLTLGNLFR SRGEVDRAI RIHQTLMESA SLTYEQRLLA IQQLGRDYMA AGLYDRAEDM FNQLTDETDF RIGALQQLLQ IYQATSEWQK A IDVAERLV ...String: MLELLFLLLP VAAAYGWYMG RRSAQQNKQD EANRLSRDYV AGVNFLLSNQ QDKAVDLFLD MLKEDTGTVE AHLTLGNLFR SRGEVDRAI RIHQTLMESA SLTYEQRLLA IQQLGRDYMA AGLYDRAEDM FNQLTDETDF RIGALQQLLQ IYQATSEWQK A IDVAERLV KLGKDKQRVE IAHFYCELAL QHMASDDLDR AMTLLKKGAA ADKNSARVSI MMGRVFMAKG EYAKAVESLQ RV ISQDREL VSETLEMLQT CYQQLGKTAE WAEFLQRAVE ENTGADAELM LADIIEARDG SEAAQVYITR QLQRHPTMRV FHK LMDYHL NEAEEGRAKE SLMVLRDMVG EKVRSKPRYR CQKCGFTAYT LYWHCPSCRA WSTIKPIRGL DGLEHHHHHH UniProtKB: Lipopolysaccharide assembly protein B |
-Macromolecule #2: Inner membrane protein YejM
| Macromolecule | Name: Inner membrane protein YejM / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.36468 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVTHRQRYRE KVSQMVSWGH WFALFNILLS LVIGSRYLFI ADWPTTLAGR IYSYVSIIGH FSFLVFATYL LILFPLTFIV GSQRLMRFL SVILATAGMT LLLIDSEVFT RFHLHLNPIV WQLVINPDEN EMARDWQLMF ISVPVILLLE LVFATWSWQK L RSLTRRRR ...String: MVTHRQRYRE KVSQMVSWGH WFALFNILLS LVIGSRYLFI ADWPTTLAGR IYSYVSIIGH FSFLVFATYL LILFPLTFIV GSQRLMRFL SVILATAGMT LLLIDSEVFT RFHLHLNPIV WQLVINPDEN EMARDWQLMF ISVPVILLLE LVFATWSWQK L RSLTRRRR FARPLAAFLF IAFIASHVVY IWADANFYRP ITMQRANLPL SYPMTARRFL EKHGLLDAQE YQRRLIEQGN PD AVSVQYP LSELRYRDMG TGQNVLLITV DGLNYSRFEK QMPALAGFAE QNISFTRHMS SGNTTDNGIF GLFYGISPSY MDG ILSTRT PAALITALNQ QGYQLGLFSS DGFTSPLYRQ ALLSDFSMPS VRTQSDEQTA TQWINWLGRY AQEDNRWFSW VSFN GTNID DSNQQAFARK YSRAAGNVDD QINRVLNALR DSGKLDNTVV IITAGRGIPL SEEEETFDWS HGHLQVPLVI HWPGT PAQR INALTDHTDL MTTLMQRLLH VSTPASEYSQ GQDLFNPQRR HYWVTAADND TLAITTPKKT LVLNNNGKYR TYNLRG ERV KDEKPQLSLL LQVLTDEKRF IAN UniProtKB: Inner membrane protein YejM |
-Macromolecule #3: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE
| Macromolecule | Name: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE type: ligand / ID: 3 / Number of copies: 1 / Formula: LPP |
|---|---|
| Molecular weight | Theoretical: 648.891 Da |
| Chemical component information |  ChemComp-LPP: |
-Macromolecule #4: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE type: ligand / ID: 4 / Number of copies: 1 / Formula: PGV |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
| Chemical component information |  ChemComp-PGV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average exposure time: 8.84 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7t6d: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)