+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human SIMC1-SLF2 complex | |||||||||
 Map data Map data | sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nse5 / Nse6 / SMC5 / SMC6 / SIMC1 / SLF2 / C5orf25 / viral restriction / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationSUMO polymer binding / positive regulation of maintenance of mitotic sister chromatid cohesion / Smc5-Smc6 complex / peptidase inhibitor activity / chromatin looping / protein localization to site of double-strand break / regulation of telomere maintenance / positive regulation of double-strand break repair / protein sumoylation / sarcomere ...SUMO polymer binding / positive regulation of maintenance of mitotic sister chromatid cohesion / Smc5-Smc6 complex / peptidase inhibitor activity / chromatin looping / protein localization to site of double-strand break / regulation of telomere maintenance / positive regulation of double-strand break repair / protein sumoylation / sarcomere / positive regulation of protein-containing complex assembly / double-strand break repair via homologous recombination / PML body / site of double-strand break / chromosome, telomeric region / DNA damage response / ubiquitin protein ligase binding / chromatin / protein-containing complex binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
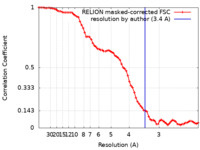
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Maeda S / Oravcova M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: The Nse5/6-like SIMC1-SLF2 complex localizes SMC5/6 to viral replication centers. Authors: Martina Oravcová / Minghua Nie / Nicola Zilio / Shintaro Maeda / Yasaman Jami-Alahmadi / Eros Lazzerini-Denchi / James A Wohlschlegel / Helle D Ulrich / Takanori Otomo / Michael N Boddy /   Abstract: The human SMC5/6 complex is a conserved guardian of genome stability and an emerging component of antiviral responses. These disparate functions likely require distinct mechanisms of SMC5/6 ...The human SMC5/6 complex is a conserved guardian of genome stability and an emerging component of antiviral responses. These disparate functions likely require distinct mechanisms of SMC5/6 regulation. In yeast, Smc5/6 is regulated by its Nse5/6 subunits, but such regulatory subunits for human SMC5/6 are poorly defined. Here, we identify a novel SMC5/6 subunit called SIMC1 that contains SUMO interacting motifs (SIMs) and an Nse5-like domain. We isolated SIMC1 from the proteomic environment of SMC5/6 within polyomavirus large T antigen (LT)-induced subnuclear compartments. SIMC1 uses its SIMs and Nse5-like domain to localize SMC5/6 to polyomavirus replication centers (PyVRCs) at SUMO-rich PML nuclear bodies. SIMC1's Nse5-like domain binds to the putative Nse6 orthologue SLF2 to form an anti-parallel helical dimer resembling the yeast Nse5/6 structure. SIMC1-SLF2 structure-based mutagenesis defines a conserved surface region containing the N-terminus of SIMC1's helical domain that regulates SMC5/6 localization to PyVRCs. Furthermore, SLF1, which recruits SMC5/6 to DNA lesions via its BRCT and ARD motifs, binds SLF2 analogously to SIMC1 and forms a separate Nse5/6-like complex. Thus, two Nse5/6-like complexes with distinct recruitment domains control human SMC5/6 localization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25706.map.gz emd_25706.map.gz | 27.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25706-v30.xml emd-25706-v30.xml emd-25706.xml emd-25706.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25706_fsc.xml emd_25706_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_25706.png emd_25706.png | 83.6 KB | ||
| Filedesc metadata |  emd-25706.cif.gz emd-25706.cif.gz | 6.1 KB | ||
| Others |  emd_25706_additional_1.map.gz emd_25706_additional_1.map.gz emd_25706_half_map_1.map.gz emd_25706_half_map_1.map.gz emd_25706_half_map_2.map.gz emd_25706_half_map_2.map.gz | 23.5 MB 23.4 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25706 http://ftp.pdbj.org/pub/emdb/structures/EMD-25706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25706 | HTTPS FTP |
-Related structure data
| Related structure data |  7t5pMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25706.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25706.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.134 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: non-sharpened
| File | emd_25706_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_25706_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_25706_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human SIMC1-SLF2 complex
| Entire | Name: Human SIMC1-SLF2 complex |
|---|---|
| Components |
|
-Supramolecule #1: Human SIMC1-SLF2 complex
| Supramolecule | Name: Human SIMC1-SLF2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SUMO-interacting motif-containing protein 1
| Macromolecule | Name: SUMO-interacting motif-containing protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.021539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AYLQDMPRSP GDVPQSPSDV SPSPDAPQSP GGMPHLPGDV LHSPGDMPHS SGDVTHSPRD IPHLPGDRPD FTQNDVQNRD MPMDISALS SPSCSPRPQS ETPLEKVPWL SVMETPARKE ISLSEPAKPG SAHVQSRTPQ GGLYNRPCLH RLKYFLRPPV H HLFFQTLI ...String: AYLQDMPRSP GDVPQSPSDV SPSPDAPQSP GGMPHLPGDV LHSPGDMPHS SGDVTHSPRD IPHLPGDRPD FTQNDVQNRD MPMDISALS SPSCSPRPQS ETPLEKVPWL SVMETPARKE ISLSEPAKPG SAHVQSRTPQ GGLYNRPCLH RLKYFLRPPV H HLFFQTLI PDKDTRENKG QRLEPIPHRR LRMVTNTIEE NFPLGTVQFL MDFVSPQHYP PREIVAHIIQ KILLSGSETV DV LKEAYML LMKIQQLHPA NAKTVEWDWK LLTYVMEEEG QTLPGRVLFL RYVVQTLEDD FQQTLRRQRQ HLQQSIANMV LSC DKQPHN VRDVIKWLVK AVTEDGLTQP PNGNQTSSGT GILKASSSHP SSQPNLTKNT NQLIVCQLQR MLSIAVEVDR TPTC SSNKI AEMMFGFVLD IPERSQREMF FTTMESHLLR CKVLEIIFLH SCETPTRLPL SLAQALYFLN NSTSLLKCQS DKSQW QTWD ELVERLQFLL SSYQHVLREH LRSSVIDRKD LIIKRIKPKP QQGDDITVVD VEKQIEAFRS RLIQMLGEPL VPQLQD KVH LLKLLLFYAA DLNPDAEPFQ KGWSGS UniProtKB: SUMO-interacting motif-containing protein 1 |
-Macromolecule #2: SMC5-SMC6 complex localization factor protein 2
| Macromolecule | Name: SMC5-SMC6 complex localization factor protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.867793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TPAATGKPPA LSKGLRSQSS DYTGHVHPGT YTNTLERLVK EMEDTQRLDE LQKQLQEDIR QGRGIKSPIR IGEEDSTDDE DGLLEEHKE FLKKFSVTID AIPDHHPGEE IFNFLNSGKI FNQYTLDLRD SGFIGQSAVE KLILKSGKTD QIFLTTQGFL T SAYHYVQC ...String: TPAATGKPPA LSKGLRSQSS DYTGHVHPGT YTNTLERLVK EMEDTQRLDE LQKQLQEDIR QGRGIKSPIR IGEEDSTDDE DGLLEEHKE FLKKFSVTID AIPDHHPGEE IFNFLNSGKI FNQYTLDLRD SGFIGQSAVE KLILKSGKTD QIFLTTQGFL T SAYHYVQC PVPVLKWLFR MMSVHTDCIV SVQILSTLME ITIRNDTFSD SPVWPWIPSL SDVAAVFFNM GIDFRSLFPL EN LQPDFNE DYLVSETQTT SRGKESEDSS YKPIFSTLPE TNILNVVKFL GLCTSIHPEG YQDREIMLLI LMLFKMSLEK QLK QIPLVD FQSLLINLMK NIRDWNTKVP ELCLGINELS SHPHNLLWLV QLVPNWTSRG RQLRQCLSLV IISKLLDEKH EDVP NASNL QVSVLHRYLV QMKPSDLLKK MVLKKKAEQP DGIIDDSLHL ELEKQAYYLT YILLHLVGEV SCSHSFSSGQ RKHFV LLCG ALEKHVKCDI REDARLFYRT KVKDLVARIH GKWQEIIQNC RPTQGQLHDF WVPDS UniProtKB: SMC5-SMC6 complex localization factor protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 88 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7t5p: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)