+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | D8-C4 computationally-designed Rotor | |||||||||||||||
 Map data Map data | D8-C4 computationally-designed rotor | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||||||||
 Authors Authors | Hansen JM / Courbet A / Quispe J / Kollman JM / Baker D | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Computational design of mechanically coupled axle-rotor protein assemblies. Authors: A Courbet / J Hansen / Y Hsia / N Bethel / Y-J Park / C Xu / A Moyer / S E Boyken / G Ueda / U Nattermann / D Nagarajan / D-A Silva / W Sheffler / J Quispe / A Nord / N King / P Bradley / D ...Authors: A Courbet / J Hansen / Y Hsia / N Bethel / Y-J Park / C Xu / A Moyer / S E Boyken / G Ueda / U Nattermann / D Nagarajan / D-A Silva / W Sheffler / J Quispe / A Nord / N King / P Bradley / D Veesler / J Kollman / D Baker /    Abstract: Natural molecular machines contain protein components that undergo motion relative to each other. Designing such mechanically constrained nanoscale protein architectures with internal degrees of ...Natural molecular machines contain protein components that undergo motion relative to each other. Designing such mechanically constrained nanoscale protein architectures with internal degrees of freedom is an outstanding challenge for computational protein design. Here we explore the de novo construction of protein machinery from designed axle and rotor components with internal cyclic or dihedral symmetry. We find that the axle-rotor systems assemble in vitro and in vivo as designed. Using cryo-electron microscopy, we find that these systems populate conformationally variable relative orientations reflecting the symmetry of the coupled components and the computationally designed interface energy landscape. These mechanical systems with internal degrees of freedom are a step toward the design of genetically encodable nanomachines. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25575.map.gz emd_25575.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25575-v30.xml emd-25575-v30.xml emd-25575.xml emd-25575.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25575.png emd_25575.png | 56.8 KB | ||
| Others |  emd_25575_additional_1.map.gz emd_25575_additional_1.map.gz emd_25575_additional_2.map.gz emd_25575_additional_2.map.gz | 242.6 KB 815.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25575 http://ftp.pdbj.org/pub/emdb/structures/EMD-25575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25575 | HTTPS FTP |
-Validation report
| Summary document |  emd_25575_validation.pdf.gz emd_25575_validation.pdf.gz | 323 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25575_full_validation.pdf.gz emd_25575_full_validation.pdf.gz | 322.6 KB | Display | |
| Data in XML |  emd_25575_validation.xml.gz emd_25575_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_25575_validation.cif.gz emd_25575_validation.cif.gz | 6.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25575 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25575 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25575.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25575.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D8-C4 computationally-designed rotor | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.32 Å | ||||||||||||||||||||||||||||||||||||
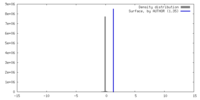
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: D8-C4 computationally-designed rotor (focused refinement on ring component)...
| File | emd_25575_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D8-C4 computationally-designed rotor (focused refinement on ring component) | ||||||||||||
| Projections & Slices |
| ||||||||||||
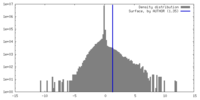
| Density Histograms |
-Additional map: D8-C4 computationally-designed rotor (focused refinement on ring component)...
| File | emd_25575_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D8-C4 computationally-designed rotor (focused refinement on ring component) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : D8-symmetric axel coupled with a C4-symmetric ring.
| Entire | Name: D8-symmetric axel coupled with a C4-symmetric ring. |
|---|---|
| Components |
|
-Supramolecule #1: D8-symmetric axel coupled with a C4-symmetric ring.
| Supramolecule | Name: D8-symmetric axel coupled with a C4-symmetric ring. / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL / In silico model: ab initio |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D4 (2x4 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 5.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 50686 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)