+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleotide-free Get3 in the closed form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tail-anchored membrane protein targeting Deviant Walker A ATPase targeting factor / CHAPERONE | |||||||||
| Biological species |  Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote) Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote) | |||||||||
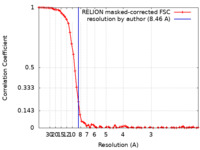
| Method | single particle reconstruction / cryo EM / Resolution: 8.46 Å | |||||||||
 Authors Authors | Fry MY / Clemons Jr WM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3. Authors: Michelle Y Fry / Vladimíra Najdrová / Ailiena O Maggiolo / Shyam M Saladi / Pavel Doležal / William M Clemons /   Abstract: Tail-anchored (TA) membrane proteins, accounting for roughly 2% of proteomes, are primarily targeted posttranslationally to the endoplasmic reticulum membrane by the guided entry of TA proteins (GET) ...Tail-anchored (TA) membrane proteins, accounting for roughly 2% of proteomes, are primarily targeted posttranslationally to the endoplasmic reticulum membrane by the guided entry of TA proteins (GET) pathway. For this complicated process, it remains unknown how the central targeting factor Get3 uses nucleotide to facilitate large conformational changes to recognize then bind clients while also preventing exposure of hydrophobic surfaces. Here, we identify the GET pathway in Giardia intestinalis and present the structure of the Get3-client complex in the critical postnucleotide-hydrolysis state, demonstrating that Get3 reorganizes the client-binding domain (CBD) to accommodate and shield the client transmembrane helix. Four additional structures of GiGet3, spanning the nucleotide-free (apo) open to closed transition and the ATP-bound state, reveal the details of nucleotide stabilization and occluded CBD. This work resolves key conundrums and allows for a complete model of the dramatic conformational landscape of Get3. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25375.map.gz emd_25375.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25375-v30.xml emd-25375-v30.xml emd-25375.xml emd-25375.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25375_fsc.xml emd_25375_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25375.png emd_25375.png | 48.8 KB | ||
| Masks |  emd_25375_msk_1.map emd_25375_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25375.cif.gz emd-25375.cif.gz | 4.9 KB | ||
| Others |  emd_25375_half_map_1.map.gz emd_25375_half_map_1.map.gz emd_25375_half_map_2.map.gz emd_25375_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25375 http://ftp.pdbj.org/pub/emdb/structures/EMD-25375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25375 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25375.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25375.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25375_msk_1.map emd_25375_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25375_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25375_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleotide-free Get3 in a closed conformation
| Entire | Name: Nucleotide-free Get3 in a closed conformation |
|---|---|
| Components |
|
-Supramolecule #1: Nucleotide-free Get3 in a closed conformation
| Supramolecule | Name: Nucleotide-free Get3 in a closed conformation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote) Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote)Strain: ATCC 50803 / WB clone C6 |
| Molecular weight | Theoretical: 78 KDa |
-Macromolecule #1: ATPase GET3
| Macromolecule | Name: ATPase GET3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote) Giardia intestinalis (strain ATCC 50803 / WB clone C6) (eukaryote) |
| Recombinant expression | Organism:  |
| Sequence | String: MLPSLHDILD QHTYKWIFFG GKGGVGKTTT SSSFSVLMAE TRPNEKFLLL STDPAHNISD AFDQKFGKAP TQVSGIPNLY AMEVDASNEM KSAVEAVQKE TGSAADNDAE SKSEGDMFGG LNDLITCASS FIKDGTFPGM DEMWSFINLI KLIDTNEYST VIFDTAPTGH ...String: MLPSLHDILD QHTYKWIFFG GKGGVGKTTT SSSFSVLMAE TRPNEKFLLL STDPAHNISD AFDQKFGKAP TQVSGIPNLY AMEVDASNEM KSAVEAVQKE TGSAADNDAE SKSEGDMFGG LNDLITCASS FIKDGTFPGM DEMWSFINLI KLIDTNEYST VIFDTAPTGH TLRFLELPET VNKVLEIFTR LKDNMGGMLS MVMQTMGLSQ NDIFGLIDKT YPKIDVVKRI SAEFRDPSLC TFVGVCIPEF LSLYETERLV QRLAVLDMDC HAIVINFVLD ANAATPCSMC RSRARMQNKY IDQINELYDD FNIVLSPLRH DEVRGIANLR DYAETLIKPY RFCWSANPDP SSAK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.55 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 9300 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Nominal magnification: 88000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)