+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2520 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
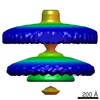
| Title | Injectisome in the tomogram of Salmonella mini-cell | |||||||||
 Map data Map data | Subtomogram averaged structure of Injectisome from Salmonella typhimurium | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Injectisome / Type III secretion system / Salmonella typhimurium / Mini-cell / Electron cryotomography | |||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 44.0 Å | |||||||||
 Authors Authors | Kawamoto A / Morimoto VY / Miyata T / Minamino T / Hughes TK / Kato T / Namba K | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2013 Journal: Sci Rep / Year: 2013Title: Common and distinct structural features of Salmonella injectisome and flagellar basal body. Authors: Akihiro Kawamoto / Yusuke V Morimoto / Tomoko Miyata / Tohru Minamino / Kelly T Hughes / Takayuki Kato / Keiichi Namba /  Abstract: Bacterial pathogens use an injectisome to deliver virulence proteins into eukaryotic host cells. The bacterial flagellum and injectisome export their component proteins for self-assembly. These two ...Bacterial pathogens use an injectisome to deliver virulence proteins into eukaryotic host cells. The bacterial flagellum and injectisome export their component proteins for self-assembly. These two systems show high structural similarities and are classified as the type III secretion system, but it remains elusive how similar they are in situ because the structures of these complexes isolated from cells and visualized by electron cryomicroscopy have shown only the export channel and housing for the export apparatus. Here we report in situ structures of Salmonella injectisome and flagellum by electron cryotomography. The injectisome lacks the flagellar basal body C-ring, but a wing-like disc and a globular density corresponding to the export gate platform and ATPase hexamer ring, respectively, are stably attached through thin connectors, revealing yet unidentified common architectures of the two systems. The ATPase ring is far from the disc, suggesting that both apparatuses are observed in an export-off state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2520.map.gz emd_2520.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2520-v30.xml emd-2520-v30.xml emd-2520.xml emd-2520.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2520-1.png EMD-2520-1.png | 91 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2520 http://ftp.pdbj.org/pub/emdb/structures/EMD-2520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2520 | HTTPS FTP |
-Validation report
| Summary document |  emd_2520_validation.pdf.gz emd_2520_validation.pdf.gz | 209.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2520_full_validation.pdf.gz emd_2520_full_validation.pdf.gz | 209.1 KB | Display | |
| Data in XML |  emd_2520_validation.xml.gz emd_2520_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2520 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2520 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2520.map.gz / Format: CCP4 / Size: 2.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2520.map.gz / Format: CCP4 / Size: 2.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram averaged structure of Injectisome from Salmonella typhimurium | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 11.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Injectisome from Salmonella typhimurium mini-cell in situ
| Entire | Name: Injectisome from Salmonella typhimurium mini-cell in situ |
|---|---|
| Components |
|
-Supramolecule #1000: Injectisome from Salmonella typhimurium mini-cell in situ
| Supramolecule | Name: Injectisome from Salmonella typhimurium mini-cell in situ type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 3.5 MDa |
-Supramolecule #1: type III secretion system
| Supramolecule | Name: type III secretion system / type: organelle_or_cellular_component / ID: 1 / Name.synonym: injectisome / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Location in cell: Plasma membrane |
| Molecular weight | Theoretical: 3.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | Details: M9 medium (17.1g Na2HPO4-12H2O, 3g KH2PO4, 0.5g NaCl, 1g NH4Cl, 0.2%glycerol, 1% typtone, 1mM MgSO4 per litre) |
|---|---|
| Grid | Details: Quantifoil molybdenum 200 mesh R0.6/1.0 grid with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80 K |
| Date | Dec 17, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON I (4k x 4k) / Average electron dose: 200 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49030 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Tilt series - Axis1 - Min angle: -70 ° / Tilt series - Axis1 - Max angle: 70 ° |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C24 (24 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 44.0 Å / Resolution method: OTHER / Software - Name: EMAN / Number subtomograms used: 75 |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















