[English] 日本語
 Yorodumi
Yorodumi- EMDB-25166: Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spike glycoprotein / antibody / VIRAL PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
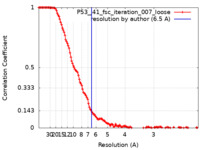
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Casner RG / Shapiro L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2022 Journal: Sci Transl Med / Year: 2022Title: An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Authors: Lihong Liu / Sho Iketani / Yicheng Guo / Eswar R Reddem / Ryan G Casner / Manoj S Nair / Jian Yu / Jasper F-W Chan / Maple Wang / Gabriele Cerutti / Zhiteng Li / Nicholas C Morano / Candace ...Authors: Lihong Liu / Sho Iketani / Yicheng Guo / Eswar R Reddem / Ryan G Casner / Manoj S Nair / Jian Yu / Jasper F-W Chan / Maple Wang / Gabriele Cerutti / Zhiteng Li / Nicholas C Morano / Candace D Castagna / Laura Corredor / Hin Chu / Shuofeng Yuan / Vincent Kwok-Man Poon / Chris Chun-Sing Chan / Zhiwei Chen / Yang Luo / Marcus Cunningham / Alejandro Chavez / Michael T Yin / David S Perlin / Moriya Tsuji / Kwok-Yung Yuen / Peter D Kwong / Zizhang Sheng / Yaoxing Huang / Lawrence Shapiro / David D Ho /   Abstract: The devastation caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has made clear the importance of pandemic preparedness. To address future zoonotic outbreaks due to related ...The devastation caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has made clear the importance of pandemic preparedness. To address future zoonotic outbreaks due to related viruses in the sarbecovirus subgenus, we identified a human monoclonal antibody, 10-40, that neutralized or bound all sarbecoviruses tested in vitro and protected against SARS-CoV-2 and SARS-CoV in vivo. Comparative studies with other receptor-binding domain (RBD)-directed antibodies showed 10-40 to have the greatest breadth against sarbecoviruses, suggesting that 10-40 is a promising agent for pandemic preparedness. Moreover, structural analyses on 10-40 and similar antibodies not only defined an epitope cluster in the inner face of the RBD that is well conserved among sarbecoviruses but also uncovered a distinct antibody class with a common CDRH3 motif. Our analyses also suggested that elicitation of this class of antibodies may not be overly difficult, an observation that bodes well for the development of a pan-sarbecovirus vaccine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25166.map.gz emd_25166.map.gz | 106.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25166-v30.xml emd-25166-v30.xml emd-25166.xml emd-25166.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25166_fsc.xml emd_25166_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25166.png emd_25166.png | 69.9 KB | ||
| Filedesc metadata |  emd-25166.cif.gz emd-25166.cif.gz | 6.1 KB | ||
| Others |  emd_25166_half_map_1.map.gz emd_25166_half_map_1.map.gz emd_25166_half_map_2.map.gz emd_25166_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25166 http://ftp.pdbj.org/pub/emdb/structures/EMD-25166 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25166 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25166 | HTTPS FTP |
-Validation report
| Summary document |  emd_25166_validation.pdf.gz emd_25166_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25166_full_validation.pdf.gz emd_25166_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_25166_validation.xml.gz emd_25166_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_25166_validation.cif.gz emd_25166_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25166 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25166 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25166 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25166 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25166.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25166.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
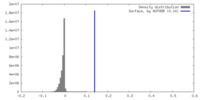
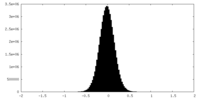
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_25166_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
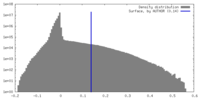
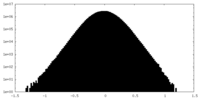
| Density Histograms |
-Half map: #1
| File | emd_25166_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
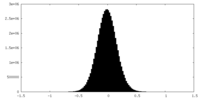
| Density Histograms |
- Sample components
Sample components
-Entire : Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike...
| Entire | Name: Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike...
| Supramolecule | Name: Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: spike glycoprotein
| Macromolecule | Name: spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRGLPQGF SALEPLVDLP IGINITRFQT LHISYLTPGD SSSGWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGNIADYNY KLPDDFTGCV IAWNSNNLDS KVGGNYNYLY RLFRKSNLKP FERDISTEIY QAGSTPCNGV KGFNCYFPLQ SYGFQPTYGV GYQPYRVVVL SFELLHAPAT VCGPKKSTNL VKNKCVNFNF NGLTGTGVLT ESNKKFLPFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ GVNCTEVPVA IHADQLTPTW RVYSTGSNVF QTRAGCLIGA EHVNNSYECD IPIGAGICAS YQTQTNSPGS ASSVASQSII AYTMSLGVEN SVAYSNNSIA IPTNFTISVT TEILPVSMTK TSVDCTMYIC GDSTECSNLL LQYGSFCTQL NRALTGIAVE QDKNTQEVFA QVKQIYKTPP IKDFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGAA LQIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN QNAQALNTLV KQLSSNFGAI SSVLNDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QSAPHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNTVYDPLQP ELDSFKEELD KYFKNHTSPD VDLGDISGIN ASVVNIQKEI DRLNEVAKNL NESLIDLQEL GKYEQ |
-Macromolecule #2: antibody 10-28 heavy
| Macromolecule | Name: antibody 10-28 heavy / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYDMHWVRQA PGKGLEWVAV ISYDGSSKFY AESVKGRFT ISRDNSKNTL YLQMNSLRAE ETAVYYCVKD GEQLVPLFDY WGQGTLVTVS S |
-Macromolecule #3: antibody 10-28 light
| Macromolecule | Name: antibody 10-28 light / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS SYLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGT DFTLTISSLQ PEDFATYYCQ QSYSTPGVTF GPGTKVDIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.4 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)