+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Androgen receptor bound to DNA - Splayed state | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
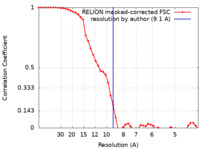
| Method | single particle reconstruction / cryo EM / Resolution: 9.1 Å | |||||||||||||||
 Authors Authors | Wasmuth EV / Vanden Broeck A / Klinge S / Sawyers CL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Allosteric interactions prime androgen receptor dimerization and activation. Authors: Elizabeth V Wasmuth / Arnaud Vanden Broeck / Justin R LaClair / Elizabeth A Hoover / Kayla E Lawrence / Navid Paknejad / Kyrie Pappas / Doreen Matthies / Biran Wang / Weiran Feng / Philip A ...Authors: Elizabeth V Wasmuth / Arnaud Vanden Broeck / Justin R LaClair / Elizabeth A Hoover / Kayla E Lawrence / Navid Paknejad / Kyrie Pappas / Doreen Matthies / Biran Wang / Weiran Feng / Philip A Watson / John C Zinder / Wouter R Karthaus / M Jason de la Cruz / Richard K Hite / Katia Manova-Todorova / Zhiheng Yu / Susan T Weintraub / Sebastian Klinge / Charles L Sawyers /  Abstract: The androgen receptor (AR) is a nuclear receptor that governs gene expression programs required for prostate development and male phenotype maintenance. Advanced prostate cancers display AR ...The androgen receptor (AR) is a nuclear receptor that governs gene expression programs required for prostate development and male phenotype maintenance. Advanced prostate cancers display AR hyperactivation and transcriptome expansion, in part, through AR amplification and interaction with oncoprotein cofactors. Despite its biological importance, how AR domains and cofactors cooperate to bind DNA has remained elusive. Using single-particle cryo-electron microscopy, we isolated three conformations of AR bound to DNA, showing that AR forms a non-obligate dimer, with the buried dimer interface utilized by ancestral steroid receptors repurposed to facilitate cooperative DNA binding. We identify novel allosteric surfaces which are compromised in androgen insensitivity syndrome and reinforced by AR's oncoprotein cofactor, ERG, and by DNA-binding motifs. Finally, we present evidence that this plastic dimer interface may have been adopted for transactivation at the expense of DNA binding. Our work highlights how fine-tuning AR's cooperative interactions translate to consequences in development and disease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25133.map.gz emd_25133.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25133-v30.xml emd-25133-v30.xml emd-25133.xml emd-25133.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25133_fsc.xml emd_25133_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25133.png emd_25133.png | 39 KB | ||
| Others |  emd_25133_half_map_1.map.gz emd_25133_half_map_1.map.gz emd_25133_half_map_2.map.gz emd_25133_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25133 http://ftp.pdbj.org/pub/emdb/structures/EMD-25133 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25133 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25133 | HTTPS FTP |
-Validation report
| Summary document |  emd_25133_validation.pdf.gz emd_25133_validation.pdf.gz | 522.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25133_full_validation.pdf.gz emd_25133_full_validation.pdf.gz | 522.2 KB | Display | |
| Data in XML |  emd_25133_validation.xml.gz emd_25133_validation.xml.gz | 10 KB | Display | |
| Data in CIF |  emd_25133_validation.cif.gz emd_25133_validation.cif.gz | 13.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25133 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25133 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25133 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25133 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25133.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25133.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.138 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_25133_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
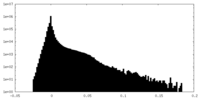
| Projections & Slices |
| ||||||||||||
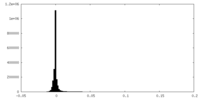
| Density Histograms |
-Half map: #2
| File | emd_25133_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
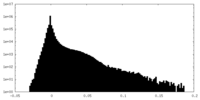
| Projections & Slices |
| ||||||||||||
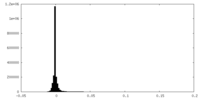
| Density Histograms |
- Sample components
Sample components
-Entire : Androgen receptor bound to DNA - Splayed state
| Entire | Name: Androgen receptor bound to DNA - Splayed state |
|---|---|
| Components |
|
-Supramolecule #1: Androgen receptor bound to DNA - Splayed state
| Supramolecule | Name: Androgen receptor bound to DNA - Splayed state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ARE DNA
| Macromolecule | Name: ARE DNA / type: dna / ID: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TAGCGTGGCC AGAACATCAT GTTCTCCGGT GCGAT |
-Macromolecule #2: Androgen receptor
| Macromolecule | Name: Androgen receptor / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: GSDYYFPPQK TCLICGDEAS GCHYGALTCG SCKVFFKRAA EGKQKYL CA SRNDCTIDKF RRKNCPSCRL RKCYEAGMTL GARKLKKLGN LKLQEEGENS NAGSPTEDPS QKMTVSHI E GYECQPIFLN VLEAIEPGVV CAGHDNNQPD SFAALLSSLN ELGERQLVHV ...String: GSDYYFPPQK TCLICGDEAS GCHYGALTCG SCKVFFKRAA EGKQKYL CA SRNDCTIDKF RRKNCPSCRL RKCYEAGMTL GARKLKKLGN LKLQEEGENS NAGSPTEDPS QKMTVSHI E GYECQPIFLN VLEAIEPGVV CAGHDNNQPD SFAALLSSLN ELGERQLVHV VKWAKALPGF RNLHVDDQM AVIQYSWMGL MVFAMGWRSF TNVNSRMLYF APDLVFNEYR MHKSRMYSQC VRMRHLSQEF GWLQITPQEF LCMKALLLF SIIPVDGLKN QKFFDELRMN YIKELDRIIA CKRKNPTSCS RRFYQLTKLL DSVQPIAREL H QFTFDLLI KSHMVSVDFP EMMAEIISVQ VPKILSGKVK PIYFHTQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)