[English] 日本語
 Yorodumi
Yorodumi- EMDB-25104: CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) T... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25104 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP in complex with Fab hVEEV-63 (focus refine of the asymmetric unit) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VLP / VEEV / viral protein / fab / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...togavirin / T=4 icosahedral viral capsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Venezuelan equine encephalitis virus (strain TC-83) / Venezuelan equine encephalitis virus (strain TC-83) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
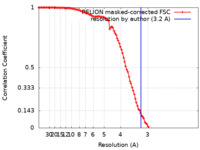
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Binshtein E / Crowe JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Exp Med / Year: 2022 Journal: J Exp Med / Year: 2022Title: Neutralizing antibodies protect mice against Venezuelan equine encephalitis virus aerosol challenge. Authors: Natasha M Kafai / Lauren E Williamson / Elad Binshtein / Soila Sukupolvi-Petty / Christina L Gardner / Jaclyn Liu / Samantha Mackin / Arthur S Kim / Nurgun Kose / Robert H Carnahan / Ana ...Authors: Natasha M Kafai / Lauren E Williamson / Elad Binshtein / Soila Sukupolvi-Petty / Christina L Gardner / Jaclyn Liu / Samantha Mackin / Arthur S Kim / Nurgun Kose / Robert H Carnahan / Ana Jung / Lindsay Droit / Douglas S Reed / Scott A Handley / William B Klimstra / James E Crowe / Michael S Diamond /  Abstract: Venezuelan equine encephalitis virus (VEEV) remains a risk for epidemic emergence or use as an aerosolized bioweapon. To develop possible countermeasures, we isolated VEEV-specific neutralizing ...Venezuelan equine encephalitis virus (VEEV) remains a risk for epidemic emergence or use as an aerosolized bioweapon. To develop possible countermeasures, we isolated VEEV-specific neutralizing monoclonal antibodies (mAbs) from mice and a human immunized with attenuated VEEV strains. Functional assays and epitope mapping established that potently inhibitory anti-VEEV mAbs bind distinct antigenic sites in the A or B domains of the E2 glycoprotein and block multiple steps in the viral replication cycle including attachment, fusion, and egress. A 3.2-Å cryo-electron microscopy reconstruction of VEEV virus-like particles bound by a human Fab suggests that antibody engagement of the B domain may result in cross-linking of neighboring spikes to prevent conformational requirements for viral fusion. Prophylaxis or postexposure therapy with these mAbs protected mice against lethal aerosol challenge with VEEV. Our study defines functional and structural mechanisms of mAb protection and suggests that multiple antigenic determinants on VEEV can be targeted for vaccine or antibody-based therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25104.map.gz emd_25104.map.gz | 63.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25104-v30.xml emd-25104-v30.xml emd-25104.xml emd-25104.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25104_fsc.xml emd_25104_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25104.png emd_25104.png | 195.7 KB | ||
| Filedesc metadata |  emd-25104.cif.gz emd-25104.cif.gz | 7.1 KB | ||
| Others |  emd_25104_half_map_1.map.gz emd_25104_half_map_1.map.gz emd_25104_half_map_2.map.gz emd_25104_half_map_2.map.gz | 80.3 MB 80.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25104 http://ftp.pdbj.org/pub/emdb/structures/EMD-25104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25104 | HTTPS FTP |
-Related structure data
| Related structure data |  7sfwMC  7sfuC  7sfvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25104.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25104.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02775 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
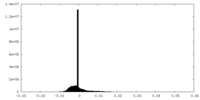
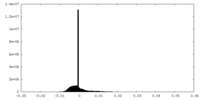
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_25104_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
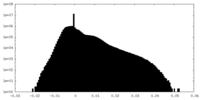
| Density Histograms |
-Half map: #2
| File | emd_25104_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
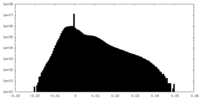
| Density Histograms |
- Sample components
Sample components
-Entire : Venezuelan equine encephalitis virus (strain TC-83)
| Entire | Name:  Venezuelan equine encephalitis virus (strain TC-83) Venezuelan equine encephalitis virus (strain TC-83) |
|---|---|
| Components |
|
-Supramolecule #1: Venezuelan equine encephalitis virus (strain TC-83)
| Supramolecule | Name: Venezuelan equine encephalitis virus (strain TC-83) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 / NCBI-ID: 11037 Sci species name: Venezuelan equine encephalitis virus (strain TC-83) Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus (strain TC-83) Venezuelan equine encephalitis virus (strain TC-83)Strain: TC-83 |
| Molecular weight | Theoretical: 47.952066 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST ...String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST AWTPFDRKIV QYAGEIYNYD FPEYGAGQPG AFGDIQSRTV SSSDLYANTN LVLQRPKAGA IHVPYTQAPS GF EQWKKDK APSLKFTAPF GCEIYTNPIR AENCAVGSIP LAFDIPDALF TRVSETPTLS AAECTLNECV YSSDFGGIAT VKY SASKSG KCAVHVPSGT ATLKEAAVEL TEQGSATIHF STANIHPEFR LQICTSYVTC KGDCHPPKDH IVTHPQYHAQ TFTA AVSKT AWTWLTSLLG GSAVIIIIGL VLATIVAMYV LTNQKHN UniProtKB: Structural polyprotein |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus (strain TC-83) Venezuelan equine encephalitis virus (strain TC-83)Strain: TC-83 |
| Molecular weight | Theoretical: 47.113746 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STEELFNEYK LTRPYMARCI RCAVGSCHSP IAIEAVKSDG HDGYVRLQTS SQYGLDSSGN LKGRTMRYDM HGTIKEIPLH QVSLYTSRP CHIVDGHGYF LLARCPAGDS ITMEFKKDSV RHSCSVPYEV KFNPVGRELY THPPEHGVEQ ACQVYAHDAQ N RGAYVEMH ...String: STEELFNEYK LTRPYMARCI RCAVGSCHSP IAIEAVKSDG HDGYVRLQTS SQYGLDSSGN LKGRTMRYDM HGTIKEIPLH QVSLYTSRP CHIVDGHGYF LLARCPAGDS ITMEFKKDSV RHSCSVPYEV KFNPVGRELY THPPEHGVEQ ACQVYAHDAQ N RGAYVEMH LPGSEVDSSL VSLSGSSVTV TPPDGTSALV ECECGGTKIS ETINKTKQFS QCTKKEQCRA YRLQNDKWVY NS DKLPKAA GATLKGKLHV PFLLADGKCT VPLAPEPMIT FGFRSVSLKL HPKNPTYLIT RQLADEPHYT HELISEPAVR NFT VTEKGW EFVWGNHPPK RFWAQETAPG NPHGLPHEVI THYYHRYPMS TILGLSICAA IATVSVAAST WLFCRSRVAC LTPY RLTPN ARIPFCLAVL CCARTARA UniProtKB: Structural polyprotein |
-Macromolecule #3: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus (strain TC-83) Venezuelan equine encephalitis virus (strain TC-83)Strain: TC-83 |
| Molecular weight | Theoretical: 30.995879 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKEA SQKQKGGGQG KKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTFP IMLEGKINGY ACVVGGKLFR PMHVEGKIDN D VLAALKTK ...String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKEA SQKQKGGGQG KKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTFP IMLEGKINGY ACVVGGKLFR PMHVEGKIDN D VLAALKTK KASKYDLEYA DVPQNMRADT FKYTHEKPQG YYSWHHGAVQ YENGRFTVPK GVGAKGDSGR PILDNQGRVV AI VLGGVNE GSRTALSVVM WNEKGVTVKY TPENCEQW UniProtKB: Structural polyprotein |
-Macromolecule #4: hVEEV-63 Fab heavy chain
| Macromolecule | Name: hVEEV-63 Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.403909 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKSSGYTFT NYIIHWVRQA PGQRLEWMGW INAGNGNTKY SQKFQGRISV TRDTSASAAY MELSSLKSE DTALYYCATL QMDYGGNGDL DYWGQGTLVT VSS |
-Macromolecule #5: hVEEV-63 Fab light chain
| Macromolecule | Name: hVEEV-63 Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.515588 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YELTQPPSVS VSPGQTARIT CSGDALPKQY VYWYQQKPGQ APVLVIYKDS ERPSGIPERF SGSSSGTTVT LTISGVQAED DADYYCQAA DSSNTEYVFG TGTKVTVL |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV / Details: Lacey Carbon. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 10586 / Average exposure time: 8.14 sec. / Average electron dose: 40.05 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7sfw: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)