+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2505 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
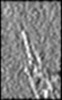
| Title | Septin-Gic1 complex in a single-tilt-axis subtomogram. | |||||||||
 Map data Map data | Cryo-Tomogram of Septin/Gic1 Complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cell division / bud neck filaments / cytokinesis / cytoskeleton / electron tomography / septin / gic | |||||||||
| Function / homology |  Function and homology information Function and homology informationseptin ring organization / Cdc42 protein signal transduction / incipient cellular bud site / cellular bud tip / regulation of exit from mitosis / cellular bud neck / mating projection tip / establishment of cell polarity / small GTPase binding / regulation of cell shape ...septin ring organization / Cdc42 protein signal transduction / incipient cellular bud site / cellular bud tip / regulation of exit from mitosis / cellular bud neck / mating projection tip / establishment of cell polarity / small GTPase binding / regulation of cell shape / cell cortex / cytoskeleton / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM / Resolution: 60.0 Å | |||||||||
 Authors Authors | Sadian Y / Gatsogiannis C / Patasi C / Hofnagel O / Goody RS / Farkasovsky M / Raunser S | |||||||||
 Citation Citation |  Journal: Elife / Year: 2013 Journal: Elife / Year: 2013Title: The role of Cdc42 and Gic1 in the regulation of septin filament formation and dissociation. Authors: Yashar Sadian / Christos Gatsogiannis / Csilla Patasi / Oliver Hofnagel / Roger S Goody / Marian Farkasovský / Stefan Raunser /  Abstract: Septins are guanine nucleotide-binding proteins that polymerize into filamentous and higher-order structures. Cdc42 and its effector Gic1 are involved in septin recruitment, ring formation and ...Septins are guanine nucleotide-binding proteins that polymerize into filamentous and higher-order structures. Cdc42 and its effector Gic1 are involved in septin recruitment, ring formation and dissociation. The regulatory mechanisms behind these processes are not well understood. Here, we have used electron microscopy and cryo electron tomography to elucidate the structural basis of the Gic1-septin and Gic1-Cdc42-septin interaction. We show that Gic1 acts as a scaffolding protein for septin filaments forming long and flexible filament cables. Cdc42 in its GTP-form binds to Gic1, which ultimately leads to the dissociation of Gic1 from the filament cables. Surprisingly, Cdc42-GDP is not inactive, but in the absence of Gic1 directly interacts with septin filaments resulting in their disassembly. We suggest that this unanticipated dual function of Cdc42 is crucial for the cell cycle. Based on our results we propose a novel regulatory mechanism for septin filament formation and dissociation. DOI: http://dx.doi.org/10.7554/eLife.01085.001. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2505.map.gz emd_2505.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2505-v30.xml emd-2505-v30.xml emd-2505.xml emd-2505.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2505.jpg emd_2505.jpg | 1.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2505 http://ftp.pdbj.org/pub/emdb/structures/EMD-2505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2505 | HTTPS FTP |
-Validation report
| Summary document |  emd_2505_validation.pdf.gz emd_2505_validation.pdf.gz | 149.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2505_full_validation.pdf.gz emd_2505_full_validation.pdf.gz | 148.2 KB | Display | |
| Data in XML |  emd_2505_validation.xml.gz emd_2505_validation.xml.gz | 4.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2505 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2505 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2505 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2505 | HTTPS FTP |
-Related structure data
| Related structure data |  2504C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2505.map.gz / Format: CCP4 / Size: 6.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2505.map.gz / Format: CCP4 / Size: 6.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-Tomogram of Septin/Gic1 Complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
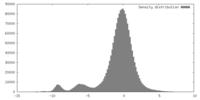
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Septin-Gic1 complex
| Entire | Name: Septin-Gic1 complex |
|---|---|
| Components |
|
-Supramolecule #1000: Septin-Gic1 complex
| Supramolecule | Name: Septin-Gic1 complex / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Macromolecule #1: Septin
| Macromolecule | Name: Septin / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Septin |
-Macromolecule #2: Gic1
| Macromolecule | Name: Gic1 / type: protein_or_peptide / ID: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: GTPase-interacting component 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 100 mM NaCl, 20 mM mM Tris-HCl, 1 mM DTT |
|---|---|
| Grid | Details: 400 Mesh 2/1 C-flat holey carbon grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 100 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Specialist optics | Energy filter - Name: in-column Omega filte / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 15.0 eV |
| Date | Oct 28, 2011 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F816 (8k x 8k) / Number real images: 63 / Average electron dose: 65 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 85470 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Tilt series - Axis1 - Min angle: -62 ° / Tilt series - Axis1 - Max angle: 62 ° / Tilt series - Axis1 - Angle increment: 2 ° |
- Image processing
Image processing
| Details | Standard eTOMO procedures for single axis tilt tomograms |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 60.0 Å / Resolution method: OTHER / Software - Name:  Imod / Number images used: 63 Imod / Number images used: 63 |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)