+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | CryoEM structure of the Caveolin-1 8S complex | |||||||||
 マップデータ マップデータ | Refined and sharpened volume of the Caveolin-1 8S complex with applied C11 symmetry | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | caveolin / caveolae / cryoEM / disc / monotopic proteins / STRUCTURAL PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報intracellular nitric oxide homeostasis / caveolar macromolecular signaling complex / protein localization to plasma membrane raft / inward rectifier potassium channel inhibitor activity / protein localization to basolateral plasma membrane / negative regulation of cytokine-mediated signaling pathway / cellular response to hyperoxia / insulin receptor internalization / caveola assembly / regulation of entry of bacterium into host cell ...intracellular nitric oxide homeostasis / caveolar macromolecular signaling complex / protein localization to plasma membrane raft / inward rectifier potassium channel inhibitor activity / protein localization to basolateral plasma membrane / negative regulation of cytokine-mediated signaling pathway / cellular response to hyperoxia / insulin receptor internalization / caveola assembly / regulation of entry of bacterium into host cell / nitric-oxide synthase inhibitor activity / positive regulation of toll-like receptor 3 signaling pathway / regulation of ruffle assembly / regulation of cardiac muscle cell action potential involved in regulation of contraction / glandular epithelial cell differentiation / negative regulation of pinocytosis / regulation of membrane repolarization during action potential / positive regulation of cell adhesion molecule production / negative regulation of potassium ion transmembrane transport / acrosomal membrane / regulation of the force of heart contraction by chemical signal / FOXO-mediated transcription of cell cycle genes / mammary gland involution / positive regulation of ERAD pathway / NOSTRIN mediated eNOS trafficking / maintenance of protein location in cell / patched binding / vesicle organization / basement membrane organization / regulation of fatty acid metabolic process / vasoconstriction / regulation of smooth muscle contraction / negative regulation of nitric oxide biosynthetic process / negative regulation of receptor signaling pathway via JAK-STAT / lipid storage / regulation of ventricular cardiac muscle cell action potential / oxysterol binding / caveolin-mediated endocytosis / cholesterol transport / negative regulation of necroptotic process / mammary gland development / protein tyrosine kinase inhibitor activity / RHOD GTPase cycle / RHOF GTPase cycle / endothelial cell proliferation / cellular response to misfolded protein / positive regulation of extrinsic apoptotic signaling pathway / Disassembly of the destruction complex and recruitment of AXIN to the membrane / RND1 GTPase cycle / RND2 GTPase cycle / RND3 GTPase cycle / Basigin interactions / cellular response to peptide hormone stimulus / peptidase activator activity / triglyceride metabolic process / cholesterol binding / RHOB GTPase cycle / post-transcriptional regulation of gene expression / muscle cell cellular homeostasis / negative regulation of epithelial cell differentiation / Triglyceride catabolism / cellular response to exogenous dsRNA / positive regulation of vasoconstriction / regulation of blood coagulation / RHOC GTPase cycle / nitric-oxide synthase binding / RHOJ GTPase cycle / negative regulation of endothelial cell proliferation / regulation of cell communication by electrical coupling involved in cardiac conduction / RHOQ GTPase cycle / regulation of heart rate by cardiac conduction / positive regulation of calcium ion transport into cytosol / CDC42 GTPase cycle / negative regulation of anoikis / positive regulation of gap junction assembly / RHOH GTPase cycle / RHOG GTPase cycle / membrane depolarization / RHOA GTPase cycle / RAC2 GTPase cycle / positive regulation of cholesterol efflux / negative regulation of BMP signaling pathway / RAC3 GTPase cycle / canonical Wnt signaling pathway / regulation of cytosolic calcium ion concentration / cellular response to transforming growth factor beta stimulus / vasculogenesis / SARS-CoV-2 targets host intracellular signalling and regulatory pathways / negative regulation of fibroblast proliferation / calcium ion homeostasis / skeletal muscle tissue development / positive regulation of intrinsic apoptotic signaling pathway / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / negative regulation of MAPK cascade / T cell costimulation / nitric oxide metabolic process / eNOS activation / lactation / receptor-mediated endocytosis of virus by host cell / lipid droplet 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
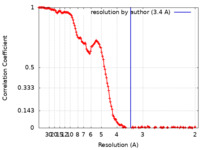
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.4 Å | |||||||||
 データ登録者 データ登録者 | Porta JP / Ohi MD | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Sci Adv / 年: 2022 ジャーナル: Sci Adv / 年: 2022タイトル: Molecular architecture of the human caveolin-1 complex. 著者: Jason C Porta / Bing Han / Alican Gulsevin / Jeong Min Chung / Yelena Peskova / Sarah Connolly / Hassane S Mchaourab / Jens Meiler / Erkan Karakas / Anne K Kenworthy / Melanie D Ohi /    要旨: Membrane-sculpting proteins shape the morphology of cell membranes and facilitate remodeling in response to physiological and environmental cues. Complexes of the monotopic membrane protein caveolin ...Membrane-sculpting proteins shape the morphology of cell membranes and facilitate remodeling in response to physiological and environmental cues. Complexes of the monotopic membrane protein caveolin function as essential curvature-generating components of caveolae, flask-shaped invaginations that sense and respond to plasma membrane tension. However, the structural basis for caveolin's membrane remodeling activity is currently unknown. Here, we show that, using cryo-electron microscopy, the human caveolin-1 complex is composed of 11 protomers organized into a tightly packed disc with a flat membrane-embedded surface. The structural insights suggest a previously unrecognized mechanism for how membrane-sculpting proteins interact with membranes and reveal how key regions of caveolin-1, including its scaffolding, oligomerization, and intramembrane domains, contribute to its function. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_25007.map.gz emd_25007.map.gz | 230 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-25007-v30.xml emd-25007-v30.xml emd-25007.xml emd-25007.xml | 15.1 KB 15.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_25007_fsc.xml emd_25007_fsc.xml | 18.3 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_25007.png emd_25007.png | 211.6 KB | ||
| Filedesc metadata |  emd-25007.cif.gz emd-25007.cif.gz | 5.9 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25007 http://ftp.pdbj.org/pub/emdb/structures/EMD-25007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25007 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_25007_validation.pdf.gz emd_25007_validation.pdf.gz | 555.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_25007_full_validation.pdf.gz emd_25007_full_validation.pdf.gz | 554.6 KB | 表示 | |
| XML形式データ |  emd_25007_validation.xml.gz emd_25007_validation.xml.gz | 14.1 KB | 表示 | |
| CIF形式データ |  emd_25007_validation.cif.gz emd_25007_validation.cif.gz | 19 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25007 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25007 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25007 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25007 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7sc0MC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_25007.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_25007.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Refined and sharpened volume of the Caveolin-1 8S complex with applied C11 symmetry | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.98 Å | ||||||||||||||||||||||||||||||||||||
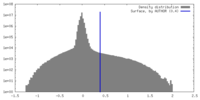
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Caveolin-1 8S complex with C11 symmetry.
| 全体 | 名称: Caveolin-1 8S complex with C11 symmetry. |
|---|---|
| 要素 |
|
-超分子 #1: Caveolin-1 8S complex with C11 symmetry.
| 超分子 | 名称: Caveolin-1 8S complex with C11 symmetry. / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Caveolin-1
| 分子 | 名称: Caveolin-1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 11 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 20.494576 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSGGKYVDSE GHLYTVPIRE QGNIYKPNNK AMADELSEKQ VYDAHTKEID LVNRDPKHLN DDVVKIDFED VIAEPEGTHS FDGIWKASF TTFTVTKYWF YRLLSALFGI PMALIWGIYF AILSFLHIWA VVPCIKSFLI EIQCISRVYS IYVHTVCDPL F EAVGKIFS NVRINLQKEI UniProtKB: Caveolin-1 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
詳細: Solutions were made fresh for each preparation of 8S particles | |||||||||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 400 / 支持フィルム - 材質: GOLD / 支持フィルム - トポロジー: HOLEY ARRAY / 支持フィルム - Film thickness: 12 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 30 sec. / 前処理 - 雰囲気: AIR / 詳細: 5 mA glow discharge | |||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 293 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS GLACIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 3838 pixel / デジタル化 - サイズ - 縦: 3710 pixel / デジタル化 - 画像ごとのフレーム数: 1-30 / 撮影したグリッド数: 1 / 実像数: 984 / 平均露光時間: 6.0 sec. / 平均電子線量: 55.5 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 100.0 µm / 最大 デフォーカス(補正後): 2.2 µm / 最小 デフォーカス(補正後): 1.5 µm / 倍率(補正後): 40103 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.2 µm / 最小 デフォーカス(公称値): 1.5 µm / 倍率(公称値): 36000 |
| 試料ステージ | 試料ホルダーモデル: OTHER / ホルダー冷却材: NITROGEN |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: AB INITIO MODEL / 温度因子: 37.8 / 当てはまり具合の基準: Correlation coefficient |
|---|---|
| 得られたモデル |  PDB-7sc0: |
 ムービー
ムービー コントローラー
コントローラー















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)