[English] 日本語
 Yorodumi
Yorodumi- EMDB-24922: Cryo-EM structure of a mammalian peptide transporter (PepT1/slc15... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
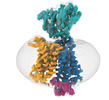
| Title | Cryo-EM structure of a mammalian peptide transporter (PepT1/slc15a1) in nanodisc | |||||||||||||||||||||
 Map data Map data | Cryo-EM map of horse PepT1 in nanodisc | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | PepT1 / SLC15 / ECD / transporter / nanodisc / TRANSPORT PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationproton-dependent oligopeptide secondary active transmembrane transporter activity / dipeptide import across plasma membrane / tripeptide transmembrane transporter activity / peptide:proton symporter activity / dipeptide transmembrane transporter activity / brush border / protein transport / apical plasma membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||||||||
 Authors Authors | Shen J / Zhou M | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Extracellular domain of PepT1 interacts with TM1 to facilitate substrate transport. Authors: Jiemin Shen / Miaohui Hu / Xiao Fan / Zhenning Ren / Corinne Portioli / Xiuwen Yan / Mingqiang Rong / Ming Zhou /  Abstract: Mammalian peptide transporters, PepT1 and PepT2, mediate uptake of small peptides and are essential for their absorption. PepT also mediates absorption of many drugs and prodrugs to enhance their ...Mammalian peptide transporters, PepT1 and PepT2, mediate uptake of small peptides and are essential for their absorption. PepT also mediates absorption of many drugs and prodrugs to enhance their bioavailability. PepT has twelve transmembrane (TM) helices that fold into an N-terminal domain (NTD, TM1-6) and a C-terminal domain (CTD, TM7-12) and has a large extracellular domain (ECD) between TM9-10. It is well recognized that peptide transport requires movements of the NTD and CTD, but the role of the ECD in PepT1 remains unclear. Here we report the structure of horse PepT1 encircled in lipid nanodiscs and captured in the inward-open apo conformation. The structure shows that the ECD bridges the NTD and CTD by interacting with TM1. Deletion of ECD or mutations to the ECD-TM1 interface impairs the transport activity. These results demonstrate an important role of ECD in PepT1 and enhance our understanding of the transport mechanism in PepT1. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24922.map.gz emd_24922.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24922-v30.xml emd-24922-v30.xml emd-24922.xml emd-24922.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24922.png emd_24922.png | 99.6 KB | ||
| Filedesc metadata |  emd-24922.cif.gz emd-24922.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24922 http://ftp.pdbj.org/pub/emdb/structures/EMD-24922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24922 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24922 | HTTPS FTP |
-Validation report
| Summary document |  emd_24922_validation.pdf.gz emd_24922_validation.pdf.gz | 425.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24922_full_validation.pdf.gz emd_24922_full_validation.pdf.gz | 425.4 KB | Display | |
| Data in XML |  emd_24922_validation.xml.gz emd_24922_validation.xml.gz | 5.4 KB | Display | |
| Data in CIF |  emd_24922_validation.cif.gz emd_24922_validation.cif.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24922 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24922 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24922 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24922 | HTTPS FTP |
-Related structure data
| Related structure data |  7s8uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24922.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24922.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of horse PepT1 in nanodisc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : PepT1 in MSP1D1 nanodisc
| Entire | Name: PepT1 in MSP1D1 nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: PepT1 in MSP1D1 nanodisc
| Supramolecule | Name: PepT1 in MSP1D1 nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 15 member 1
| Macromolecule | Name: Solute carrier family 15 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.723992 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGMSRSQSCF GYPLSIFFIV VNEFCERFSY YGMRALLILY FTLFIGWDDN LSTAIYHTFV ALCYLTPILG ALIADSWLGK FKTIVSLSI VYTIGQVVLA VSSINDLTDG NRDGTPDSLP VHVALSMIGL ALIAFGTGGI KPCVSAFGGD QFEEGQEKQR N RFFSIFYL ...String: MGMSRSQSCF GYPLSIFFIV VNEFCERFSY YGMRALLILY FTLFIGWDDN LSTAIYHTFV ALCYLTPILG ALIADSWLGK FKTIVSLSI VYTIGQVVLA VSSINDLTDG NRDGTPDSLP VHVALSMIGL ALIAFGTGGI KPCVSAFGGD QFEEGQEKQR N RFFSIFYL AINAGSLLST IITPILRVQQ CGIHSKQACY PLAFGVPAAL MAVSLIVFVI GSRMYKKFQP QGNIMAKVAK CI GFAITNR IRHRSNKFPK REHWLDWAKE KYDERLISQI KMVTRVMFLY IPLPMFWALF DQQGSRWTLQ ATTMNGQIGV IEI QPDQMQ TVNAILIVIM VPIMDAVVYP LIAKCGLNFT SLKKMTVGMF LASMAFVAAA IVQVEIDKTL PVFPNGHEVQ VKVL NIGNN SMNISFPGET MAVNQMSQTG DFMTFDVDKL SINISSTGSP VTPVTHNFEQ GHRHTILVWA PNHYRVIKDG LDRKP EKGQ NGIRFVNAFD KSFDVTMDGT VYVNVTSHSA SEYQFFPSGK KSFTINSTEI SQQCERNFTS PRLGFGSAYT YVMGRK ADG CPELSVFGDI PPNTVNMALQ IPQYFLLTCG EVVFSVTGLE FSYSQAPSNM KSVLQAGWLL TVAVGNIIVL IVAGAGQ FS KQWAEYILFA ALLLVVCVIF GIMAQFYTYV NPAEVEAKFD DDEKKKNPEK ENPYYTLDSV SQTRM UniProtKB: Solute carrier family 15 member 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.9000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















