+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GLP-1 receptor bound with Pfizer small molecule agonist | |||||||||
 Map data Map data | EM density map for GLP1R | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GLP-1R / GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucagon-like peptide 1 receptor activity / glucagon receptor activity / positive regulation of blood pressure / hormone secretion / post-translational protein targeting to membrane, translocation / response to psychosocial stress / regulation of heart contraction / peptide hormone binding / activation of adenylate cyclase activity / negative regulation of blood pressure ...glucagon-like peptide 1 receptor activity / glucagon receptor activity / positive regulation of blood pressure / hormone secretion / post-translational protein targeting to membrane, translocation / response to psychosocial stress / regulation of heart contraction / peptide hormone binding / activation of adenylate cyclase activity / negative regulation of blood pressure / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon-type ligand receptors / transmembrane signaling receptor activity / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events / learning or memory / cell surface receptor signaling pathway / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
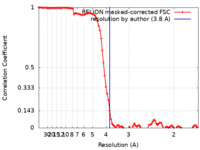
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Liu Y / Dias JM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2022 Journal: J Med Chem / Year: 2022Title: A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor. Authors: David A Griffith / David J Edmonds / Jean-Philippe Fortin / Amit S Kalgutkar / J Brent Kuzmiski / Paula M Loria / Aditi R Saxena / Scott W Bagley / Clare Buckeridge / John M Curto / David R ...Authors: David A Griffith / David J Edmonds / Jean-Philippe Fortin / Amit S Kalgutkar / J Brent Kuzmiski / Paula M Loria / Aditi R Saxena / Scott W Bagley / Clare Buckeridge / John M Curto / David R Derksen / João M Dias / Matthew C Griffor / Seungil Han / V Margaret Jackson / Margaret S Landis / Daniel Lettiere / Chris Limberakis / Yuhang Liu / Alan M Mathiowetz / Jayesh C Patel / David W Piotrowski / David A Price / Roger B Ruggeri / David A Tess /   Abstract: Peptide agonists of the glucagon-like peptide-1 receptor (GLP-1R) have revolutionized diabetes therapy, but their use has been limited because they require injection. Herein, we describe the ...Peptide agonists of the glucagon-like peptide-1 receptor (GLP-1R) have revolutionized diabetes therapy, but their use has been limited because they require injection. Herein, we describe the discovery of the orally bioavailable, small-molecule, GLP-1R agonist PF-06882961 (danuglipron). A sensitized high-throughput screen was used to identify 5-fluoropyrimidine-based GLP-1R agonists that were optimized to promote endogenous GLP-1R signaling with nanomolar potency. Incorporation of a carboxylic acid moiety provided considerable GLP-1R potency gains with improved off-target pharmacology and reduced metabolic clearance, ultimately resulting in the identification of danuglipron. Danuglipron increased insulin levels in primates but not rodents, which was explained by receptor mutagensis studies and a cryogenic electron microscope structure that revealed a binding pocket requiring a primate-specific tryptophan 33 residue. Oral administration of danuglipron to healthy humans produced dose-proportional increases in systemic exposure (NCT03309241). This opens an opportunity for oral small-molecule therapies that target the well-validated GLP-1R for metabolic health. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24794.map.gz emd_24794.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24794-v30.xml emd-24794-v30.xml emd-24794.xml emd-24794.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24794_fsc.xml emd_24794_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_24794.png emd_24794.png | 64.8 KB | ||
| Filedesc metadata |  emd-24794.cif.gz emd-24794.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24794 http://ftp.pdbj.org/pub/emdb/structures/EMD-24794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24794 | HTTPS FTP |
-Related structure data
| Related structure data |  7s15MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24794.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24794.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM density map for GLP1R | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.848 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : agonist bound GLP1 receptor
| Entire | Name: agonist bound GLP1 receptor |
|---|---|
| Components |
|
-Supramolecule #1: agonist bound GLP1 receptor
| Supramolecule | Name: agonist bound GLP1 receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: GLP1 receptor is stabilized by StaR mutations |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glucagon-like peptide 1 receptor
| Macromolecule | Name: Glucagon-like peptide 1 receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.701004 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RPQGATVSLW ETVQKWREYR RQCQRSLTED PPPATDLFCN RTFDEYACWP DGEPGSFVNV SCPWYLPWAS SVPQGHVYRF CTAEGLWLQ KDNSSLPWRD LSECEESKRG ERSSPEEQLL FLYYIYTVGY ALSFSALVIA SAILLGFRHL HCTRNYIHLN L FASFILRA ...String: RPQGATVSLW ETVQKWREYR RQCQRSLTED PPPATDLFCN RTFDEYACWP DGEPGSFVNV SCPWYLPWAS SVPQGHVYRF CTAEGLWLQ KDNSSLPWRD LSECEESKRG ERSSPEEQLL FLYYIYTVGY ALSFSALVIA SAILLGFRHL HCTRNYIHLN L FASFILRA LSVFIKDAAL KWMYSTRAQQ HEWDGLLRYQ DSLSCRLVFL LMQYCVAANY YWLLVEGVYL YTLLAFSVAS EQ WIFRLYV SIGWGVPLLF VVPWGIVKYL AEDEGCWTRN SNMNYWLIIR LPILFAIGVN FLIFVRVICI VVSKLKANEM CKT DIQCRL AKSTLTLIPL LGTHEVIFAF VMDEHARGTL RFIKLFTELS FTSFQGLMVA ILYCFVNNEV QLEFRKSWER WRL UniProtKB: Glucagon-like peptide 1 receptor |
-Macromolecule #2: 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl...
| Macromolecule | Name: 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid type: ligand / ID: 2 / Number of copies: 1 / Formula: 82L |
|---|---|
| Molecular weight | Theoretical: 586.632 Da |
| Chemical component information | 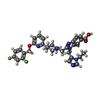 ChemComp-82L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.075 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II / Details: -2 blotting force 4S blotting time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-20 / Number real images: 19000 / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)