+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24468 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of apo-state of human CNGA3/CNGB3 channel | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cone / CNG channel / cGMP / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinorganic cation import across plasma membrane / intracellular cyclic nucleotide activated cation channel complex / intracellularly cGMP-activated cation channel activity / intracellularly cAMP-activated cation channel activity / transmembrane transporter complex / axon initial segment / ligand-gated monoatomic ion channel activity / myosin binding / sodium channel activity / monoatomic cation transmembrane transport ...inorganic cation import across plasma membrane / intracellular cyclic nucleotide activated cation channel complex / intracellularly cGMP-activated cation channel activity / intracellularly cAMP-activated cation channel activity / transmembrane transporter complex / axon initial segment / ligand-gated monoatomic ion channel activity / myosin binding / sodium channel activity / monoatomic cation transmembrane transport / cGMP binding / response to magnesium ion / glial cell projection / monoatomic cation transport / photoreceptor outer segment / response to cAMP / visual perception / calcium channel activity / perikaryon / cadherin binding / dendrite / protein-containing complex binding / signal transduction / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Zheng X / Yang J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure of the human cone photoreceptor cyclic nucleotide-gated channel. Authors: Xiangdong Zheng / Zhengshan Hu / Huan Li / Jian Yang /  Abstract: Cyclic nucleotide-gated (CNG) channels transduce light-induced chemical signals into electrical signals in retinal cone and rod photoreceptors. Structures of native CNG channels, which are ...Cyclic nucleotide-gated (CNG) channels transduce light-induced chemical signals into electrical signals in retinal cone and rod photoreceptors. Structures of native CNG channels, which are heterotetramers formed by CNGA and CNGB subunits, have not been obtained. In the present study, we report a high-resolution cryo-electron microscopy structure of the human cone CNG channel in the apo closed state. The channel contains three CNGA3 and one CNGB3 subunits. Arg403 in the pore helix of CNGB3 projects into an asymmetric selectivity filter and forms hydrogen bonds with two pore-lining backbone carbonyl oxygens. Arg442 in S6 of CNGB3 protrudes into and occludes the pore below the hydrophobic cavity gate previously observed in homotetrameric CNGA channels. It is interesting that Arg403Gln is a disease mutation, and Arg442 is replaced by glutamine in some animal species with dichromatic or monochromatic vision. These and other unique structural features and the disease link conferred by CNGB3 indicate a critical role of CNGB3 in shaping cone photoresponses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24468.map.gz emd_24468.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24468-v30.xml emd-24468-v30.xml emd-24468.xml emd-24468.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24468.png emd_24468.png | 164.1 KB | ||
| Filedesc metadata |  emd-24468.cif.gz emd-24468.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24468 http://ftp.pdbj.org/pub/emdb/structures/EMD-24468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24468 | HTTPS FTP |
-Validation report
| Summary document |  emd_24468_validation.pdf.gz emd_24468_validation.pdf.gz | 511.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24468_full_validation.pdf.gz emd_24468_full_validation.pdf.gz | 511.4 KB | Display | |
| Data in XML |  emd_24468_validation.xml.gz emd_24468_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_24468_validation.cif.gz emd_24468_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24468 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24468 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24468 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24468 | HTTPS FTP |
-Related structure data
| Related structure data |  7rhsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24468.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24468.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8247 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
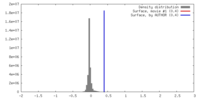
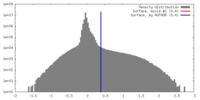
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : native human cone photoreceptor heterotetrameric CNG channel CNGA...
| Entire | Name: native human cone photoreceptor heterotetrameric CNG channel CNGA3/CNGB3 |
|---|---|
| Components |
|
-Supramolecule #1: native human cone photoreceptor heterotetrameric CNG channel CNGA...
| Supramolecule | Name: native human cone photoreceptor heterotetrameric CNG channel CNGA3/CNGB3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cyclic nucleotide-gated cation channel alpha-3
| Macromolecule | Name: Cyclic nucleotide-gated cation channel alpha-3 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.233195 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGSMAKINT QYSHPSRTHL KVKTSDRDLN RAENGLSRAH SSSEETSSVL QPGIAMETRG LADSGQGSFT GQGIARLSRL IFLLRRWAA RHVHHQDQGP DSFPDRFRGA ELKEVSSQES NAQANVGSQE PADRGRSAWP LAKCNTNTSN NTEEEKKTKK K DAIVVDPS ...String: GPGSMAKINT QYSHPSRTHL KVKTSDRDLN RAENGLSRAH SSSEETSSVL QPGIAMETRG LADSGQGSFT GQGIARLSRL IFLLRRWAA RHVHHQDQGP DSFPDRFRGA ELKEVSSQES NAQANVGSQE PADRGRSAWP LAKCNTNTSN NTEEEKKTKK K DAIVVDPS SNLYYRWLTA IALPVFYNWY LLICRACFDE LQSEYLMLWL VLDYSADVLY VLDVLVRART GFLEQGLMVS DT NRLWQHY KTTTQFKLDV LSLVPTDLAY LKVGTNYPEV RFNRLLKFSR LFEFFDRTET RTNYPNMFRI GNLVLYILII IHW NACIYF AISKFIGFGT DSWVYPNISI PEHGRLSRKY IYSLYWSTLT LTTIGETPPP VKDEEYLFVV VDFLVGVLIF ATIV GNVGS MISNMNASRA EFQAKIDSIK QYMQFRKVTK DLETRVIRWF DYLWANKKTV DEKEVLKSLP DKLKAEIAIN VHLDT LKKV RIFQDCEAGL LVELVLKLRP TVFSPGDYIC KKGDIGKEMY IINEGKLAVV ADDGVTQFVV LSDGSYFGEI SILNIK GSK SGNRRTANIR SIGYSDLFCL SKDDLMEALT EYPEAKKALE EKGRQILMKD NLIDEELARA GADPKDLEEK VEQLGSS LD TLQTRFARLL AEYNATQMKM KQRLSQLESQ VKGGGDKPLA DGEVPGDATK TEDKQQDYKD DDDK UniProtKB: Cyclic nucleotide-gated channel alpha-3 |
-Macromolecule #2: Cyclic nucleotide-gated cation channel beta-3
| Macromolecule | Name: Cyclic nucleotide-gated cation channel beta-3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 95.450758 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSWSHPQFE KGGGSGGGSG GSAWSHPQFE KGSFKSLTKV NKVKPIGENN ENEQSSRRNE EGSHPSNQSQ QTTAQEENKG EEKSLKTKS TPVTSEEPHT NIQDKLSKKN SSGDLTTNPD PQNAAEPTGT VPEQKEMDPG KEGPNSPQNK PPAAPVINEY A DAQLHNLV ...String: MGSWSHPQFE KGGGSGGGSG GSAWSHPQFE KGSFKSLTKV NKVKPIGENN ENEQSSRRNE EGSHPSNQSQ QTTAQEENKG EEKSLKTKS TPVTSEEPHT NIQDKLSKKN SSGDLTTNPD PQNAAEPTGT VPEQKEMDPG KEGPNSPQNK PPAAPVINEY A DAQLHNLV KRMRQRTALY KKKLVEGDLS SPEASPQTAK PTAVPPVKES DDKPTEHYYR LLWFKVKKMP LTEYLKRIKL PN SIDSYTD RLYLLWLLLV TLAYNWNCCF IPLRLVFPYQ TADNIHYWLI ADIICDIIYL YDMLFIQPRL QFVRGGDIIV DSN ELRKHY RTSTKFQLDV ASIIPFDICY LFFGFNPMFR ANRMLKYTSF FEFNHHLESI MDKAYIYRVI RTTGYLLFIL HINA CVYYW ASNYEGIGTT RWVYDGEGNE YLRCYYWAVR TLITIGGLPE PQTLFEIVFQ LLNFFSGVFV FSSLIGQMRD VIGAA TANQ NYFRACMDDT IAYMNNYSIP KLVQKRVRTW YEYTWDSQRM LDESDLLKTL PTTVQLALAI DVNFSIISKV DLFKGC DTQ MIYDMLLRLK SVLYLPGDFV CKKGEIGKEM YIIKHGEVQV LGGPDGTKVL VTLKAGSVFG EISLLAAGGG NRRTANV VA HGFANLLTLD KKTLQEILVH YPDSERILMK KARVLLKQKA KTAEATPPRK DLALLFPPKE ETPKLFKTLL GGTGKASL A RLLKLKREQA AQKKENSEGG EEEGKENEDK QKENEDKQKE NEDKGKENED KDKGREPEEK PLDRPECTAS PIAVEEEPH SVRRTVLPRG TSRQSLIISM APSAEGGEEV LTIEVKEKAK Q UniProtKB: Cyclic nucleotide-gated channel beta-3 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 8.58 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 57.63 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)