+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Spike in complex with PVI.V6-14 Fab | |||||||||
 Map data Map data | Locally refined, sharpened map of SARS-CoV-2 Spike NTD in complex with PVI.V6-14 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / neutralizing antibody / vaccine / plasmablast / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationimmunoglobulin complex / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...immunoglobulin complex / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / adaptive immune response / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / extracellular region / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
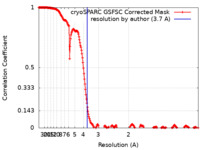
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Altomare CG / Bajic G | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2022 Journal: mBio / Year: 2022Title: Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal Domain. Authors: Clara G Altomare / Daniel C Adelsberg / Juan Manuel Carreno / Iden A Sapse / Fatima Amanat / Ali H Ellebedy / Viviana Simon / Florian Krammer / Goran Bajic /  Abstract: Structural characterization of infection- and vaccination-elicited antibodies in complex with antigen provides insight into the evolutionary arms race between the host and the pathogen and informs ...Structural characterization of infection- and vaccination-elicited antibodies in complex with antigen provides insight into the evolutionary arms race between the host and the pathogen and informs rational vaccine immunogen design. We isolated a germ line-encoded monoclonal antibody (mAb) from plasmablasts activated upon mRNA vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and determined its structure in complex with the spike glycoprotein by electron cryomicroscopy (cryo-EM). We show that the mAb engages a previously uncharacterized neutralizing epitope on the spike N-terminal domain (NTD). The high-resolution structure reveals details of the intermolecular interactions and shows that the mAb inserts its heavy complementarity-determining region 3 (HCDR3) loop into a hydrophobic NTD cavity previously shown to bind a heme metabolite, biliverdin. We demonstrate direct competition with biliverdin and that, because of the conserved nature of the epitope, the mAb maintains binding to viral variants B.1.1.7 (alpha), B.1.351 (beta), B.1.617.2 (delta), and B.1.1.529 (omicron). Our study describes a novel conserved epitope on the NTD that is readily targeted by vaccine-induced antibody responses. We report the first structure of a vaccine-induced antibody to SARS-CoV-2 spike isolated from plasmablasts 7 days after vaccination. The genetic sequence of the antibody PVI.V6-14 suggests that it is completely unmutated, meaning that this type of B cell did not undergo somatic hypermutation or affinity maturation; this cell was likely already present in the donor and was activated by the vaccine. This is, to our knowledge, also the first structure of an unmutated antibody in complex with its cognate antigen. PVI.V6-14 binds a novel, conserved epitope on the N-terminal domain (NTD) and neutralizes the original viral strain. PVI.V6-14 also binds the newly emerged variants B.1.1.7 (alpha), B.1.351 (beta), B.1.617.2 (delta), and B.1.1.529 (omicron). Given that this antibody was likely already present in the donor prior to vaccination, we believe that this antibody class could potentially "keep up" with the new variants, should they continue to emerge, by undergoing somatic hypermutation and affinity maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24402.map.gz emd_24402.map.gz | 902.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24402-v30.xml emd-24402-v30.xml emd-24402.xml emd-24402.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24402_fsc.xml emd_24402_fsc.xml | 21.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_24402.png emd_24402.png | 25.9 KB | ||
| Filedesc metadata |  emd-24402.cif.gz emd-24402.cif.gz | 6.4 KB | ||
| Others |  emd_24402_additional_1.map.gz emd_24402_additional_1.map.gz | 512.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24402 http://ftp.pdbj.org/pub/emdb/structures/EMD-24402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24402 | HTTPS FTP |
-Related structure data
| Related structure data |  7rbuMC  7rbvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11127 (Title: Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal Domain EMPIAR-11127 (Title: Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal DomainData size: 5.1 TB Data #1: Unaligned multi-frame movies of SARS-CoV-2 spike glycoprotein in complex with PVI.V6-14 Fab [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24402.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24402.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined, sharpened map of SARS-CoV-2 Spike NTD in complex with PVI.V6-14 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.56 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Locally refined, unsharpened map of SARS-CoV-2 Spike NTD...
| File | emd_24402_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined, unsharpened map of SARS-CoV-2 Spike NTD in complex with PVI.V6-14 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Spike:PVI.V6-14 Fab complex
| Entire | Name: SARS-CoV-2 Spike:PVI.V6-14 Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Spike:PVI.V6-14 Fab complex
| Supramolecule | Name: SARS-CoV-2 Spike:PVI.V6-14 Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 550 kDa/nm |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.417703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFT UniProtKB: Spike glycoprotein |
-Macromolecule #2: PVI.V6-14 Fab heavy chain, variable region
| Macromolecule | Name: PVI.V6-14 Fab heavy chain, variable region / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.932396 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCTVSGGSIS SSSYYWGWIR QPPGKGLEWI GSIYYSGSTY YNPSLKSRVT ISVDTSKNQF SLKLSSVTA ADTAVYYCAR CRPEYYFGSG SYLDFDYWGQ GTLVTVSS |
-Macromolecule #3: PVI.V6-14 Fab light chain, variable region
| Macromolecule | Name: PVI.V6-14 Fab light chain, variable region / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.385669 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS VSASVGDRVT ITCRASQGIS SWLAWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ QANSFPLTFG GGTKVEIK UniProtKB: IGL c1310_light_IGKV1-12_IGKJ4 |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)