+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24291 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-ET platelet of control mouse (1 week) | |||||||||
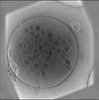
 Map data Map data | Representative tomographic reconstruction of platelet from control mouse(1 week). Bin4. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Huo T / Wang Z | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Using Cryo-ET to distinguish platelets during pre-acute myeloid leukemia from steady state hematopoiesis. Authors: Yuewei Wang / Tong Huo / Yu-Jung Tseng / Lan Dang / Zhili Yu / Wenjuan Yu / Zachary Foulks / Rebecca L Murdaugh / Steven J Ludtke / Daisuke Nakada / Zhao Wang /   Abstract: Early diagnosis of acute myeloid leukemia (AML) in the pre-leukemic stage remains a clinical challenge, as pre-leukemic patients show no symptoms, lacking any known morphological or numerical ...Early diagnosis of acute myeloid leukemia (AML) in the pre-leukemic stage remains a clinical challenge, as pre-leukemic patients show no symptoms, lacking any known morphological or numerical abnormalities in blood cells. Here, we demonstrate that platelets with structurally abnormal mitochondria emerge at the pre-leukemic phase of AML, preceding detectable changes in blood cell counts or detection of leukemic blasts in blood. We visualized frozen-hydrated platelets from mice at different time points during AML development in situ using electron cryo-tomography (cryo-ET) and identified intracellular organelles through an unbiased semi-automatic process followed by quantitative measurement. A large proportion of platelets exhibited changes in the overall shape and depletion of organelles in AML. Notably, 23% of platelets in pre-leukemic cells exhibit abnormal, round mitochondria with unfolded cristae, accompanied by a significant drop in ATP levels and altered expression of metabolism-related gene signatures. Our study demonstrates that detectable structural changes in pre-leukemic platelets may serve as a biomarker for the early diagnosis of AML. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24291.map.gz emd_24291.map.gz | 1.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24291-v30.xml emd-24291-v30.xml emd-24291.xml emd-24291.xml | 7.4 KB 7.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24291.png emd_24291.png | 184.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24291 http://ftp.pdbj.org/pub/emdb/structures/EMD-24291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24291 | HTTPS FTP |
-Validation report
| Summary document |  emd_24291_validation.pdf.gz emd_24291_validation.pdf.gz | 234 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24291_full_validation.pdf.gz emd_24291_full_validation.pdf.gz | 233.5 KB | Display | |
| Data in XML |  emd_24291_validation.xml.gz emd_24291_validation.xml.gz | 4.6 KB | Display | |
| Data in CIF |  emd_24291_validation.cif.gz emd_24291_validation.cif.gz | 5.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24291 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24291 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24291.map.gz / Format: CCP4 / Size: 2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24291.map.gz / Format: CCP4 / Size: 2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Representative tomographic reconstruction of platelet from control mouse(1 week). Bin4. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 48.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Platelet of control mouse (1 week)
| Entire | Name: Platelet of control mouse (1 week) |
|---|---|
| Components |
|
-Supramolecule #1: Platelet of control mouse (1 week)
| Supramolecule | Name: Platelet of control mouse (1 week) / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: AURION / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 31 |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)

















