[English] 日本語
 Yorodumi
Yorodumi- EMDB-24284: Azoarcus group I intron, symmetry-expanded monomer from a synthet... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Azoarcus group I intron, symmetry-expanded monomer from a synthetic trimeric construct | ||||||||||||||||||
 Map data Map data | Azoarcus group I intron, symmetry-expanded monomer from a synthetic trimeric construct | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Azoarcus olearius (bacteria) Azoarcus olearius (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | ||||||||||||||||||
 Authors Authors | Thelot F / Liu D / Liao M / Yin P | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2022 Journal: Nat Methods / Year: 2022Title: Sub-3-Å cryo-EM structure of RNA enabled by engineered homomeric self-assembly. Authors: Di Liu / François A Thélot / Joseph A Piccirilli / Maofu Liao / Peng Yin /  Abstract: High-resolution structural studies are essential for understanding the folding and function of diverse RNAs. Herein, we present a nanoarchitectural engineering strategy for efficient structural ...High-resolution structural studies are essential for understanding the folding and function of diverse RNAs. Herein, we present a nanoarchitectural engineering strategy for efficient structural determination of RNA-only structures using single-particle cryogenic electron microscopy (cryo-EM). This strategy-ROCK (RNA oligomerization-enabled cryo-EM via installing kissing loops)-involves installing kissing-loop sequences onto the functionally nonessential stems of RNAs for homomeric self-assembly into closed rings with multiplied molecular weights and mitigated structural flexibility. ROCK enables cryo-EM reconstruction of the Tetrahymena group I intron at 2.98-Å resolution overall (2.85 Å for the core), allowing de novo model building of the complete RNA, including the previously unknown peripheral domains. ROCK is further applied to two smaller RNAs-the Azoarcus group I intron and the FMN riboswitch, revealing the conformational change of the former and the bound ligand in the latter. ROCK holds promise to greatly facilitate the use of cryo-EM in RNA structural studies. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24284.map.gz emd_24284.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24284-v30.xml emd-24284-v30.xml emd-24284.xml emd-24284.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24284.png emd_24284.png | 64.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24284 http://ftp.pdbj.org/pub/emdb/structures/EMD-24284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24284 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24284.map.gz / Format: CCP4 / Size: 5.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24284.map.gz / Format: CCP4 / Size: 5.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Azoarcus group I intron, symmetry-expanded monomer from a synthetic trimeric construct | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||
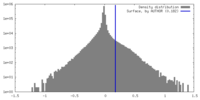
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Azoarcus group I intron, symmetry-expanded monomer from a synthet...
| Entire | Name: Azoarcus group I intron, symmetry-expanded monomer from a synthetic trimeric construct |
|---|---|
| Components |
|
-Supramolecule #1: Azoarcus group I intron, symmetry-expanded monomer from a synthet...
| Supramolecule | Name: Azoarcus group I intron, symmetry-expanded monomer from a synthetic trimeric construct type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Azoarcus olearius (bacteria) Azoarcus olearius (bacteria) |
| Molecular weight | Theoretical: 80 KDa |
-Macromolecule #1: Azoarcus group I intron
| Macromolecule | Name: Azoarcus group I intron / type: rna / ID: 1 |
|---|---|
| Source (natural) | Organism:  Azoarcus olearius (bacteria) Azoarcus olearius (bacteria) |
| Sequence | String: GGCCGUGUGC CUUGCGCCGG GAAACCACGC AAGGGAUGGU GUCAAAUUCG GCGAAACCUA AGCGCGGAUG GUUCGCGUAU GGCAACGCCG AGCCAAGCUU CGCAGCCUUC GGGCUGCGAU GAAGGUGUAG AGACUAGACG GCACCCACCU AAGGCGCACC GAACCAUCCG ...String: GGCCGUGUGC CUUGCGCCGG GAAACCACGC AAGGGAUGGU GUCAAAUUCG GCGAAACCUA AGCGCGGAUG GUUCGCGUAU GGCAACGCCG AGCCAAGCUU CGCAGCCUUC GGGCUGCGAU GAAGGUGUAG AGACUAGACG GCACCCACCU AAGGCGCACC GAACCAUCCG GUGCGCUAUG GUGAAGGCAU AGUCCAGGGA GUGGCGAAAG CCACACAAAC CAG |
-Macromolecule #2: Ligated exon mimic of the Azoarcus Group I intron
| Macromolecule | Name: Ligated exon mimic of the Azoarcus Group I intron / type: other / ID: 2 Classification: polydeoxyribonucleotide/polyribonucleotide hybrid |
|---|---|
| Source (natural) | Organism:  Azoarcus olearius (bacteria) Azoarcus olearius (bacteria) |
| Sequence | String: CATACGGCC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.0 mm / Nominal magnification: 31000 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 486860 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)