+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of commercially purchased Apoferritin | |||||||||
 Map data Map data | Cryo-EM map of Apoferritin purchased commercially | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.91 Å | |||||||||
 Authors Authors | Moser TJ / Parvate AD / Evans JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2022 Journal: Front Mol Biosci / Year: 2022Title: Cryo-EM structure of the diapause chaperone artemin. Authors: Amar D Parvate / Samantha M Powell / Jory T Brookreson / Trevor H Moser / Irina V Novikova / Mowei Zhou / James E Evans /  Abstract: The protein artemin acts as both an RNA and protein chaperone and constitutes over 10% of all protein in cysts during diapause. However, its mechanistic details remain elusive since no high- ...The protein artemin acts as both an RNA and protein chaperone and constitutes over 10% of all protein in cysts during diapause. However, its mechanistic details remain elusive since no high-resolution structure of artemin exists. Here we report the full-length structure of artemin at 2.04 Å resolution. The cryo-EM map contains density for an intramolecular disulfide bond between Cys22-Cys61 and resolves the entire C-terminus extending into the core of the assembled protein cage but in a different configuration than previously hypothesized with molecular modeling. We also provide data supporting the role of C-terminal helix F towards stabilizing the dimer form that is believed to be important for its chaperoning activity. We were able to destabilize this effect by placing a tag at the C-terminus to fully pack the internal cavity and cause limited steric hindrance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24145.map.gz emd_24145.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24145-v30.xml emd-24145-v30.xml emd-24145.xml emd-24145.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24145.png emd_24145.png | 53.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24145 http://ftp.pdbj.org/pub/emdb/structures/EMD-24145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24145 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24145.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24145.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Apoferritin purchased commercially | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.4108 Å | ||||||||||||||||||||||||||||||||||||
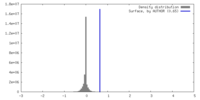
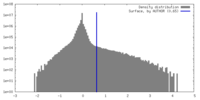
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Apoferritin
| Entire | Name: Apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Apoferritin
| Supramolecule | Name: Apoferritin / type: complex / ID: 1 / Parent: 0 Details: Commercially purchased fully assembled protein complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 444 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 Details: Solutions made fresh and filtered with 0.22 micron filter for sterility. |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: LEICA EM GP / Details: 2 s blot. |
| Details | Sample was monodisperse and suspended in TBS |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: OTHER / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 11028 / Average exposure time: 0.5 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -1.3 µm / Nominal defocus min: -0.3 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)