[English] 日本語
 Yorodumi
Yorodumi- EMDB-24065: CryoEM map of monoclonal antibody Fab DH1025.1 bound to bound to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of monoclonal antibody Fab DH1025.1 bound to bound to CH505.M5 SOSIP trimer | |||||||||
 Map data Map data | CryoEM map of DH1025.1 bound to CH505.M5 SOSIP trimer | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Manne K / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: HIV-1 vaccine induction of protective tier 2 neutralizing antibodies in nonhuman primates is augmented by TLR7/8 agonism Authors: Tilahun K / Haynes BF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24065.map.gz emd_24065.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24065-v30.xml emd-24065-v30.xml emd-24065.xml emd-24065.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24065.png emd_24065.png | 34.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24065 http://ftp.pdbj.org/pub/emdb/structures/EMD-24065 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24065 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24065 | HTTPS FTP |
-Related structure data
| Related structure data |  7mw9 M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24065.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24065.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of DH1025.1 bound to CH505.M5 SOSIP trimer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
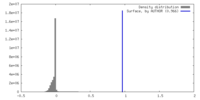
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : DH1025.1 Fab bound to CH505.M5 SOSIP trimer
| Entire | Name: DH1025.1 Fab bound to CH505.M5 SOSIP trimer |
|---|---|
| Components |
|
-Supramolecule #1: DH1025.1 Fab bound to CH505.M5 SOSIP trimer
| Supramolecule | Name: DH1025.1 Fab bound to CH505.M5 SOSIP trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F Homo sapiens (human) / Recombinant cell: 293F |
-Macromolecule #1: CH505.M5 gp120
| Macromolecule | Name: CH505.M5 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 50.965219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ENLWVTVYYG VPVWKEAKTT LFCASDAKAY EKEVHNVWAT HACVPTDPNP QEMVLKNVTE NFNMWKNDMV DQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS V ITQACPKV ...String: ENLWVTVYYG VPVWKEAKTT LFCASDAKAY EKEVHNVWAT HACVPTDPNP QEMVLKNVTE NFNMWKNDMV DQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS V ITQACPKV SFDPIPIHYC APAGYAILKC NNKTFTGTGP CNNVSTVQCT HGIKPVVSTQ LLLNGSLAEG EIIIRSENIT KN VKTIIVH LNESVKIECT RPNNKTRTSI RIGPGQAFYA TGQVIGDIRE AYCNINESKW NETLQRVSKK LKEYFPHKNI TFQ PSSGGD LEITTHSFNC GGEFFYCNTS SLFNRTYMAN NSTRTITIHC RIKQIINMWQ EVGRAMYAPP IAGNITCISN ITGL LLTRD YGKNNTETFR PGGGNMKDNW RSELYKYKVV KIEPLGVAPT RCKRRVV |
-Macromolecule #2: CH505.M5 gp41
| Macromolecule | Name: CH505.M5 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 11.238608 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VWGIKQLQAR VLAVERYLRD QQLLGIWGCS GKLICCTNVP WNSSWSNRNL SEIWDNMTWL QWDKEISNYT QIIYGLLEES QNQQEKNEQ DLLALD |
-Macromolecule #3: DH1025.1 Fab Heavy Chain
| Macromolecule | Name: DH1025.1 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.238191 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHLQESGPG LVKPSETLSL TCAVSGGSVS YYNWWSWIRQ PPGKGLEWIG YISGSSGNTY YSPSLKSRVT ISTDTSKNQF SLRLSSVTA ADTAVYYCAS VLQYLDWVLP DRVGEVHWFD VWGPGVLVSV SSASTKGPSV FPLAPSSRST SESTAALGCL V KDYFPEPV ...String: QVHLQESGPG LVKPSETLSL TCAVSGGSVS YYNWWSWIRQ PPGKGLEWIG YISGSSGNTY YSPSLKSRVT ISTDTSKNQF SLRLSSVTA ADTAVYYCAS VLQYLDWVLP DRVGEVHWFD VWGPGVLVSV SSASTKGPSV FPLAPSSRST SESTAALGCL V KDYFPEPV TVSWNSGSLT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYVCNVNH KPSNTKVDKR VEIKTCGG |
-Macromolecule #4: DH1025.1 Fab Light Chain
| Macromolecule | Name: DH1025.1 Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.12957 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ISCQASQGIS SRLAWYQQRP GKAPKLLIHT ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QHHSHPLTFG GGTKVEIKRA VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGVLKTGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDRVT ISCQASQGIS SRLAWYQQRP GKAPKLLIHT ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QHHSHPLTFG GGTKVEIKRA VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGVLKTGNS Q ESVTEQDS KDNTYSLSST LTLSNTDYQS HNVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 293 K / Instrument: LEICA EM GP / Details: Blot for 2.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.15 K / Max: 93.15 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3056 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model | 
PDB-7mw9: |
 Movie
Movie Controller
Controller


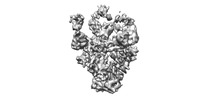
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)