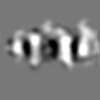[English] 日本語
 Yorodumi
Yorodumi- EMDB-23791: Subtomogram average of Photosystem II 2D semicrystalline array on... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23791 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of Photosystem II 2D semicrystalline array on thylakoid membranes isolated from Phaeodactylum tricornutum | |||||||||
 Map data Map data | Subtomogram average of Photosystem II 2D semicrystalline array on thylakoid membranes isolated from Phaeodactylum tricornutum | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 37.0 Å | |||||||||
 Authors Authors | Jiang J / Cheong KY / Falkowski PG / Dai W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2021 Journal: J Struct Biol / Year: 2021Title: Integrating on-grid immunogold labeling and cryo-electron tomography to reveal photosystem II structure and spatial distribution in thylakoid membranes. Authors: Jennifer Jiang / Kuan Yu Cheong / Paul G Falkowski / Wei Dai /  Abstract: A long-standing challenge in cell biology is elucidating the structure and spatial distribution of individual membrane-bound proteins, protein complexes and their interactions in their native ...A long-standing challenge in cell biology is elucidating the structure and spatial distribution of individual membrane-bound proteins, protein complexes and their interactions in their native environment. Here, we describe a workflow that combines on-grid immunogold labeling, followed by cryo-electron tomography (cryoET) imaging and structural analyses to identify and characterize the structure of photosystem II (PSII) complexes. Using an antibody specific to a core subunit of PSII, the D1 protein (uniquely found in the water splitting complex in all oxygenic photoautotrophs), we identified PSII complexes in biophysically active thylakoid membranes isolated from a model marine diatom Phaeodactylum tricornutum. Subsequent cryoET analyses of these protein complexes resolved two PSII structures: supercomplexes and dimeric cores. Our integrative approach establishes the structural signature of multimeric membrane protein complexes in their native environment and provides a pathway to elucidate their high-resolution structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23791.map.gz emd_23791.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23791-v30.xml emd-23791-v30.xml emd-23791.xml emd-23791.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23791.png emd_23791.png | 100.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23791 http://ftp.pdbj.org/pub/emdb/structures/EMD-23791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23791 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23791.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23791.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of Photosystem II 2D semicrystalline array on thylakoid membranes isolated from Phaeodactylum tricornutum | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.454 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Photosystem II 2D semicrystalline array
| Entire | Name: Photosystem II 2D semicrystalline array |
|---|---|
| Components |
|
-Supramolecule #1: Photosystem II 2D semicrystalline array
| Supramolecule | Name: Photosystem II 2D semicrystalline array / type: complex / ID: 1 / Parent: 0 Details: On thylakoid membranes isolated from Phaeodactylum tricornutum |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 49000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 37.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN2 / Number subtomograms used: 263 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 263 |
| CTF correction | Software - Name: EMAN2 |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)