+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E1435Q Ycf1 mutant in inward-facing narrow conformation | |||||||||
 Map data Map data | Sharpened refine map constructed on unsharpened map using provided mask by Sharpen 3D - cisTEM. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type Cd2+ transporter / ABC-type cadmium transporter activity / Recycling of bile acids and salts / Heme degradation / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / P-type cadmium transporter activity / bilirubin transmembrane transporter activity / bilirubin transport ...ABC-type Cd2+ transporter / ABC-type cadmium transporter activity / Recycling of bile acids and salts / Heme degradation / Aspirin ADME / Atorvastatin ADME / Paracetamol ADME / P-type cadmium transporter activity / bilirubin transmembrane transporter activity / bilirubin transport / ABC-family proteins mediated transport / vacuole fusion, non-autophagic / ABC-type glutathione S-conjugate transporter activity / ABC-type glutathione-S-conjugate transporter / fungal-type vacuole / fungal-type vacuole membrane / response to metal ion / ATPase-coupled transmembrane transporter activity / response to cadmium ion / glutathione metabolic process / cell redox homeostasis / transmembrane transport / membrane raft / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
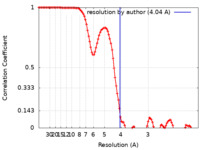
| Method | single particle reconstruction / cryo EM / Resolution: 4.04 Å | |||||||||
 Authors Authors | Khandelwal NK / Millan CR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: The structural basis for regulation of the glutathione transporter Ycf1 by regulatory domain phosphorylation. Authors: Nitesh Kumar Khandelwal / Cinthia R Millan / Samantha I Zangari / Samantha Avila / Dewight Williams / Tarjani M Thaker / Thomas M Tomasiak /  Abstract: Yeast Cadmium Factor 1 (Ycf1) sequesters heavy metals and glutathione into the vacuole to counter cell stress. Ycf1 belongs to the ATP binding cassette C-subfamily (ABCC) of transporters, many of ...Yeast Cadmium Factor 1 (Ycf1) sequesters heavy metals and glutathione into the vacuole to counter cell stress. Ycf1 belongs to the ATP binding cassette C-subfamily (ABCC) of transporters, many of which are regulated by phosphorylation on intrinsically-disordered domains. The regulatory mechanism of phosphorylation is still poorly understood. Here, we report two cryo-EM structures of Ycf1 at 3.4 Å and 4.0 Å resolution in inward-facing open conformations that capture previously unobserved ordered states of the intrinsically disordered regulatory domain (R-domain). R-domain phosphorylation is clearly evident and induces a topology promoting electrostatic and hydrophobic interactions with Nucleotide Binding Domain 1 (NBD1) and the Lasso motif. These interactions stay constant between the structures and are related by rigid body movements of the NBD1/R-domain complex. Biochemical data further show R-domain phosphorylation reorganizes the Ycf1 architecture and is required for maximal ATPase activity. Together, we provide insights into how R-domains control ABCC transporter activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23690.map.gz emd_23690.map.gz | 95.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23690-v30.xml emd-23690-v30.xml emd-23690.xml emd-23690.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23690_fsc.xml emd_23690_fsc.xml | 12.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23690.png emd_23690.png | 39.4 KB | ||
| Masks |  emd_23690_msk_1.map emd_23690_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23690.cif.gz emd-23690.cif.gz | 7.4 KB | ||
| Others |  emd_23690_additional_1.map.gz emd_23690_additional_1.map.gz emd_23690_half_map_1.map.gz emd_23690_half_map_1.map.gz emd_23690_half_map_2.map.gz emd_23690_half_map_2.map.gz | 95.4 MB 33.8 MB 33.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23690 http://ftp.pdbj.org/pub/emdb/structures/EMD-23690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23690 | HTTPS FTP |
-Related structure data
| Related structure data |  7m68MC  7m69C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23690.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23690.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened refine map constructed on unsharpened map using provided mask by Sharpen 3D - cisTEM. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.031 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_23690_msk_1.map emd_23690_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
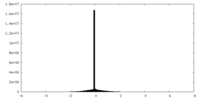
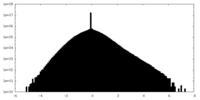
| Density Histograms |
-Additional map: Unsharpened map obtained from Manual Refine - cisTEM.
| File | emd_23690_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map obtained from Manual Refine - cisTEM. | ||||||||||||
| Projections & Slices |
| ||||||||||||
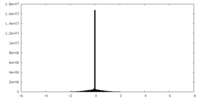
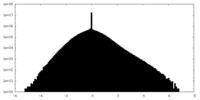
| Density Histograms |
-Half map: The second half-map from unsharpened refine map generated...
| File | emd_23690_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second half-map from unsharpened refine map generated in Generate 3D - cisTEM. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The first half-map from unsharpened refine map generated...
| File | emd_23690_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first half-map from unsharpened refine map generated in Generate 3D - cisTEM. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ycf1 monomer
| Entire | Name: Ycf1 monomer |
|---|---|
| Components |
|
-Supramolecule #1: Ycf1 monomer
| Supramolecule | Name: Ycf1 monomer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 176.66831 KDa |
-Macromolecule #1: Metal resistance protein YCF1
| Macromolecule | Name: Metal resistance protein YCF1 / type: protein_or_peptide / ID: 1 Details: Phosphorylated residues (S908, T911 and S914) are present in structure file. Number of copies: 1 / Enantiomer: LEVO / EC number: ABC-type Cd2+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 176.318328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASDYKDDDDK GALEVLFQGP SSPMAGNLVS WACKLCRSPE GFGPISFYGD FTQCFIDGVI LNLSAIFMIT FGIRDLVNLC KKKHSGIKY RRNWIIVSRM ALVLLEIAFV SLASLNISKE EAENFTIVSQ YASTMLSLFV ALALHWIEYD RSVVANTVLL F YWLFETFG ...String: ASDYKDDDDK GALEVLFQGP SSPMAGNLVS WACKLCRSPE GFGPISFYGD FTQCFIDGVI LNLSAIFMIT FGIRDLVNLC KKKHSGIKY RRNWIIVSRM ALVLLEIAFV SLASLNISKE EAENFTIVSQ YASTMLSLFV ALALHWIEYD RSVVANTVLL F YWLFETFG NFAKLINILI RHTYEGIWYS GQTGFILTLF QVITCASILL LEALPKKPLM PHQHIHQTLT RRKPNPYDSA NI FSRITFS WMSGLMKTGY EKYLVEADLY KLPRNFSSEE LSQKLEKNWE NELKQKSNPS LSWAICRTFG SKMLLAAFFK AIH DVLAFT QPQLLRILIK FVTDYNSERQ DDHSSLQGFE NNHPQKLPIV RGFLIAFAMF LVGFTQTSVL HQYFLNVFNT GMYI KSALT ALIYQKSLVL SNEASGLSST GDIVNLMSVD VQKLQDLTQW LNLIWSGPFQ IIICLYSLYK LLGNSMWVGV IILVI MMPL NSFLMRIQKK LQKSQMKYKD ERTRVISEIL NNIKSLKLYA WEKPYREKLE EVRNNKELKN LTKLGCYMAV TSFQFN IVP FLVSCCTFAV FVYTEDRALT TDLVFPALTL FNLLSFPLMI IPMVLNSFIE ASVSIGRLFT FFTNEELQPD SVQRLPK VK NIGDVAINIG DDATFLWQRK PEYKVALKNI NFQAKKGNLT CIVGKVGSGK TALLSCMLGD LFRVKGFATV HGSVAYVS Q VPWIMNGTVK ENILFGHRYD AEFYEKTIKA CALTIDLAIL MDGDKTLVGE KGISLSGGQK ARLSLARAVY ARADTYLLD DPLAAVDEHV ARHLIEHVLG PNGLLHTKTK VLATNKVSAL SIADSIALLD NGEITQQGTY DEITKDADSP LWKLLNNYGK KNNGKSNEF GDSSESSVRE SSIPVEGELE QLQKLNDLDF GNSDAISLRR A(SEP)DA(TPO)LG(SEP)ID FGDDENIAK REHREQGKVK WNIYLEYAKA CNPKSVCVFI LFIVISMFLS VMGNVWLKHW SEVNSRYGSN PNAARYLAIY FALGIGSALA TLIQTIVLW VFCTIHASKY LHNLMTNSVL RAPMTFFETT PIGRILNRFS NDIYKVDALL GRTFSQFFVN AVKVTFTITV I CATTWQFI FIIIPLSVFY IYYQQYYLRT SRELRRLDSI TRSPIYSHFQ ETLGGLATVR GYSQQKRFSH INQCRIDNNM SA FYPSINA NRWLAYRLEL IGSIIILGAA TLSVFRLKQG TLTAGMVGLS LSYALQITQT LNWIVRMTVE VETNIVSVER IKE YADLKS EAPLIVEGHR PPKEWPSQGD IKFNNYSTRY RPELDLVLKH INIHIKPNEK VGIVGRTGAG KSSLTLALFR MIEA SEGNI VIDNIAINEI GLYDLRHKLS IIPQDSQVFE GTVRENIDPI NQYTDEAIWR ALELSHLKEH VLSMSNDGLD AQLTE GGGN LSVGQRQLLC LARAMLVPSK ILVLDQATAA VDVETDKVVQ ETIRTAFKDR TILTIAHRLN TIMDSDRIIV LDNGKV AEF DSPGQLLSDN KSLFYSLCME AGLVNENGLV PRGSSAHHHH HHHHHHGA UniProtKB: Metal resistance protein YCF1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.94 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: Solution were made fresh in cold distilled water and final pH was adjusted to 7.0 with HCl of cold buffer. The digitonin detergent was added to final .06 % in buffer after pH adjustment. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: Pelco 15mA - 30s glow/30s hold charge | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 8499 / Average exposure time: 2.9 sec. / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)