+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23625 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
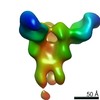
| Title | HMPV F v3-B in complex with MPE33 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | |||||||||
| Biological species |  Human metapneumovirus / Human metapneumovirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.79 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses. Authors: Guillaume B E Stewart-Jones / Jason Gorman / Li Ou / Baoshan Zhang / M Gordon Joyce / Lijuan Yang / Cheng Cheng / Gwo-Yu Chuang / Kathryn E Foulds / Wing-Pui Kong / Adam S Olia / Mallika ...Authors: Guillaume B E Stewart-Jones / Jason Gorman / Li Ou / Baoshan Zhang / M Gordon Joyce / Lijuan Yang / Cheng Cheng / Gwo-Yu Chuang / Kathryn E Foulds / Wing-Pui Kong / Adam S Olia / Mallika Sastry / Chen-Hsiang Shen / John-Paul Todd / Yaroslav Tsybovsky / Raffaello Verardi / Yongping Yang / Peter L Collins / Davide Corti / Antonio Lanzavecchia / Diana G Scorpio / John R Mascola / Ursula J Buchholz / Peter D Kwong /   Abstract: Human metapneumovirus (HMPV) is a major cause of respiratory disease worldwide, particularly among children and the elderly. Although there is no licensed HMPV vaccine, promising candidates have been ...Human metapneumovirus (HMPV) is a major cause of respiratory disease worldwide, particularly among children and the elderly. Although there is no licensed HMPV vaccine, promising candidates have been identified for related pneumoviruses based on the structure-based stabilization of the fusion (F) glycoprotein trimer, with prefusion-stabilized F glycoprotein trimers eliciting significantly higher neutralizing responses than their postfusion F counterparts. However, immunization with HMPV F trimers in either prefusion or postfusion conformations has been reported to elicit equivalent neutralization responses. Here we investigate the impact of stabilizing disulfides, especially interprotomer disulfides (IP-DSs) linking protomers of the F trimer, on the elicitation of HMPV-neutralizing responses. We designed F trimer disulfides, screened for their expression, and used electron microscopy (EM) to confirm their formation, including that of an unexpected postfusion variant. In mice, IP-DS-stabilized prefusion and postfusion HMPV F elicited significantly higher neutralizing responses than non-IP-DS-stabilized HMPV Fs. In macaques, the impact of IP-DS stabilization was more measured, although IP-DS-stabilized variants of either prefusion or postfusion HMPV F induced neutralizing responses many times the average titers observed in a healthy human cohort. Serological and absorption-based analyses of macaque responses revealed elicited HMPV-neutralizing responses to be absorbed differently by IP-DS-containing and by non-IP-DS-containing postfusion Fs, suggesting IP-DS stabilization to alter not only the immunogenicity of select epitopes but their antigenicity as well. We speculate the observed increase in immunogenicity by IP-DS trimers to be related to reduced interprotomer flexibility within the HMPV F trimer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23625.map.gz emd_23625.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23625-v30.xml emd-23625-v30.xml emd-23625.xml emd-23625.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23625.png emd_23625.png | 94 KB | ||
| Masks |  emd_23625_msk_1.map emd_23625_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_23625_additional_1.map.gz emd_23625_additional_1.map.gz emd_23625_additional_2.map.gz emd_23625_additional_2.map.gz emd_23625_half_map_1.map.gz emd_23625_half_map_1.map.gz emd_23625_half_map_2.map.gz emd_23625_half_map_2.map.gz | 120 MB 8.5 MB 226.4 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23625 http://ftp.pdbj.org/pub/emdb/structures/EMD-23625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23625 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23625.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23625.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23625_msk_1.map emd_23625_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
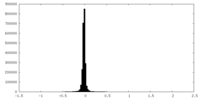
| Density Histograms |
-Additional map: unsharpened map
| File | emd_23625_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
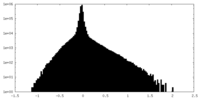
| Density Histograms |
-Additional map: density modified map with ResolveCryoEM
| File | emd_23625_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified map with ResolveCryoEM | ||||||||||||
| Projections & Slices |
| ||||||||||||
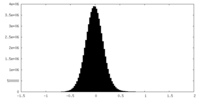
| Density Histograms |
-Half map: half map B
| File | emd_23625_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_23625_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HMPV F v3-B in complex with MPE33
| Entire | Name: HMPV F v3-B in complex with MPE33 |
|---|---|
| Components |
|
-Supramolecule #1: HMPV F v3-B in complex with MPE33
| Supramolecule | Name: HMPV F v3-B in complex with MPE33 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: HMPV F v3-B
| Supramolecule | Name: HMPV F v3-B / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: MPE33
| Supramolecule | Name: MPE33 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: HMPV F v3-B
| Macromolecule | Name: HMPV F v3-B / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TPQHGLKESY LEESCSTITE GYLSVLRTGW YTNVFTLEVG DVENLTCTDG PSLIKTELDL TKSALRELKT CSADQLAREE QIEGGGGGGF VLGAIALGVA TAAAVTAGIA IAKTIRLESE VNAIKGCLKT TNECVSTLGN GVRVLATAVR ELKEFVSKNL TSAINKNKCD ...String: TPQHGLKESY LEESCSTITE GYLSVLRTGW YTNVFTLEVG DVENLTCTDG PSLIKTELDL TKSALRELKT CSADQLAREE QIEGGGGGGF VLGAIALGVA TAAAVTAGIA IAKTIRLESE VNAIKGCLKT TNECVSTLGN GVRVLATAVR ELKEFVSKNL TSAINKNKCD IPDLKMAVSF SQFNRRFLNV VRQFSDNAGI TPAISLDLMT DAELARAVSY MPTSAGQIKL MLENRCMVRR KGFGILIGVY GSSVIYMVQL PIFGVIDTPC WIIKAAPSCS EKDGNYACLL REDQGWYCKN AGSTVYYPND KDCETRGDHV FCDTAAGINV AEQSRECNIN ISTTNYPCKV STGRHPISMV ALSPLGALVA CYKGVSCSIG SNRVGIIKQL PKGCSYITNQ DADTVTIDNT VYQLSKVEGE QHVIKGRPVS SSFDPIKFPE DQFNVALDQV FESIENSQAL VDQSNKILNS AESAIGGYIP EAPRDGQAYV RKDGEWVLLS TFLGGLVPR |
-Macromolecule #2: MPE33 Heavy chain
| Macromolecule | Name: MPE33 Heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: tgvhsqvqlq esgpglvkps qtlsltctvs gdsisngsyc wnwvrqpagk glewigriyp igntnynpsl ksrvtisldt sknqfslkla svtaadtavy ycarearife gyyyyyygld vwgqgttvtv ssastkgpsv fplapsskst sggtaalgcl vkdyfpepvt ...String: tgvhsqvqlq esgpglvkps qtlsltctvs gdsisngsyc wnwvrqpagk glewigriyp igntnynpsl ksrvtisldt sknqfslkla svtaadtavy ycarearife gyyyyyygld vwgqgttvtv ssastkgpsv fplapsskst sggtaalgcl vkdyfpepvt vswnsgalts gvhtfpavlq ssglyslssv vtvpssslgt qtyicnvnhk psntkvdkkv epkscdkglv pr |
-Macromolecule #3: MPE33 Light chain
| Macromolecule | Name: MPE33 Light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPAT LSLSPGERAT LSCRASQTIS SYLAWYQQKP GQAPRLLISD VSKRATGIPA RFSGSGSGTD FTLTISSLEP EDFAVYYCHQ RSNWDTFGQG TKLEIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 63.78 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller







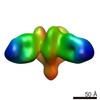





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)