[English] 日本語
 Yorodumi
Yorodumi- EMDB-23462: Structure of CD4 mimetic BNM-III-170 in complex with BG505 SOSIP.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23462 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CD4 mimetic BNM-III-170 in complex with BG505 SOSIP.664 HIV-1 Env trimer and 17b Fab | |||||||||
 Map data Map data | Map of BNM-III-170-BG505-17b Fab complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CD4m / fusion / HIV-1 / entry / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Jette CA / Bjorkman PJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structures of HIV-1 trimer bound to CD4-mimetics BNM-III-170 and M48U1 adopt a CD4-bound open conformation. Authors: Claudia A Jette / Christopher O Barnes / Sharon M Kirk / Bruno Melillo / Amos B Smith / Pamela J Bjorkman /  Abstract: Human immunodeficiency virus-1 (HIV-1), the causative agent of AIDS, impacts millions of people. Entry into target cells is mediated by the HIV-1 envelope (Env) glycoprotein interacting with host ...Human immunodeficiency virus-1 (HIV-1), the causative agent of AIDS, impacts millions of people. Entry into target cells is mediated by the HIV-1 envelope (Env) glycoprotein interacting with host receptor CD4, which triggers conformational changes allowing binding to a coreceptor and subsequent membrane fusion. Small molecule or peptide CD4-mimetic drugs mimic CD4's Phe43 interaction with Env by inserting into the conserved Phe43 pocket on Env subunit gp120. Here, we present single-particle cryo-EM structures of CD4-mimetics BNM-III-170 and M48U1 bound to a BG505 native-like Env trimer plus the CD4-induced antibody 17b at 3.7 Å and 3.9 Å resolution, respectively. CD4-mimetic-bound BG505 exhibits canonical CD4-induced conformational changes including trimer opening, formation of the 4-stranded gp120 bridging sheet, displacement of the V1V2 loop, and formation of a compact and elongated gp41 HR1C helical bundle. We conclude that CD4-induced structural changes on both gp120 and gp41 Env subunits are induced by binding to the gp120 Phe43 pocket. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23462.map.gz emd_23462.map.gz | 5.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23462-v30.xml emd-23462-v30.xml emd-23462.xml emd-23462.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23462.png emd_23462.png | 52.4 KB | ||
| Filedesc metadata |  emd-23462.cif.gz emd-23462.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23462 http://ftp.pdbj.org/pub/emdb/structures/EMD-23462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23462 | HTTPS FTP |
-Related structure data
| Related structure data |  7lo6MC  7lokC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23462.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23462.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of BNM-III-170-BG505-17b Fab complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.104 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of BG505 SOSIP.664 Env with CD4 mimetic BNM-III-170 and 1...
| Entire | Name: Complex of BG505 SOSIP.664 Env with CD4 mimetic BNM-III-170 and 17b Fab |
|---|---|
| Components |
|
-Supramolecule #1: Complex of BG505 SOSIP.664 Env with CD4 mimetic BNM-III-170 and 1...
| Supramolecule | Name: Complex of BG505 SOSIP.664 Env with CD4 mimetic BNM-III-170 and 17b Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: 17b Fab
| Supramolecule | Name: 17b Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3-#4 Details: Final reconstruction masked out the constant domain so only coordinates for the variable domain were modeled, but full Fab was used to assemble complex. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: BG505 SOSIP.664 HIV-1 Env trimer
| Supramolecule | Name: BG505 SOSIP.664 HIV-1 Env trimer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
-Macromolecule #1: Envelope glycoprotein BG505 SOSIP.664 gp120
| Macromolecule | Name: Envelope glycoprotein BG505 SOSIP.664 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.864086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLWVTVYYGV PVWKDAETTL FCASDAKAYE TEKHNVWATH ACVPTDPNPQ EIHLENVTEE FNMWKNNMVE QMHTDIISLW DQSLKPCVK LTPLCVTLQC TNVTNNITDD MRGELKNCSF NMTTELRDKK QKVYSLFYRL DVVQINENQG NRSNNSNKEY R LINCNTSA ...String: NLWVTVYYGV PVWKDAETTL FCASDAKAYE TEKHNVWATH ACVPTDPNPQ EIHLENVTEE FNMWKNNMVE QMHTDIISLW DQSLKPCVK LTPLCVTLQC TNVTNNITDD MRGELKNCSF NMTTELRDKK QKVYSLFYRL DVVQINENQG NRSNNSNKEY R LINCNTSA ITQACPKVSF EPIPIHYCAP AGFAILKCKD KKFNGTGPCP SVSTVQCTHG IKPVVSTQLL LNGSLAEEEV MI RSENITN NAKNILVQFN TPVQINCTRP NNNTRKSIRI GPGQAFYATG DIIGDIRQAH CNVSKATWNE TLGKVVKQLR KHF GNNTII RFANSSGGDL EVTTHSFNCG GEFFYCNTSG LFNSTWISNT SVQGSNSTGS NDSITLPCRI KQIINMWQRI GQAM YAPPI QGVIRCVSNI TGLILTRDGG STNSTTETFR PGGGDMRDNW RSELYKYKVV KIEPLGVAPT RCKRRVVGRR RRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41
| Macromolecule | Name: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: 17b Fab Light Chain
| Macromolecule | Name: 17b Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.399898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPAT LSVSPGERAT LSCRASESVS SDLAWYQQKP GQAPRLLIYG ASTRATGVPA RFSGSGSGAE FTLTISSLQS EDFAVYYCQ QYNNWPPRYT FGQGTRLEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: DIVMTQSPAT LSVSPGERAT LSCRASESVS SDLAWYQQKP GQAPRLLIYG ASTRATGVPA RFSGSGSGAE FTLTISSLQS EDFAVYYCQ QYNNWPPRYT FGQGTRLEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRG |
-Macromolecule #4: 17b Fab Heavy Chain
| Macromolecule | Name: 17b Fab Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.748771 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGAE VKKPGSSVKV SCKASGDTFI RYSFTWVRQA PGQGLEWMGR IITILDVAHY APHLQGRVTI TADKSTSTVY LELRNLRSD DTAVYFCAGV YEGEADEGEY DNNGFLKHWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS ...String: EVQLVESGAE VKKPGSSVKV SCKASGDTFI RYSFTWVRQA PGQGLEWMGR IITILDVAHY APHLQGRVTI TADKSTSTVY LELRNLRSD DTAVYFCAGV YEGEADEGEY DNNGFLKHWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKRVEP KSCDKHHHHH H |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #10: ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminometh...
| Macromolecule | Name: ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide type: ligand / ID: 10 / Number of copies: 3 / Formula: 5VG |
|---|---|
| Molecular weight | Theoretical: 446.906 Da |
| Chemical component information | 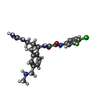 ChemComp-5VG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.5 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 662861 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-7lo6: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)