+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23378 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Mayaro virus | ||||||||||||||||||
 Map data Map data | CryoEM density map of MAYV focused region | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | alphavirus / togavirus / enveloped virus / pathogen / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Mayaro virus Mayaro virus | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||||||||
 Authors Authors | Chmielewski D / Kaelber J | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Near-atomic resolution Cryo-EM structure of Mayaro virus identifies key structural determinants of alphavirus particle formation Authors: Chmielewski D / Kaelber JT / Jin J / Weaver S / Auguste AJ / Chiu W | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23378.map.gz emd_23378.map.gz | 205.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23378-v30.xml emd-23378-v30.xml emd-23378.xml emd-23378.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23378.png emd_23378.png | 239.7 KB | ||
| Filedesc metadata |  emd-23378.cif.gz emd-23378.cif.gz | 5.8 KB | ||
| Others |  emd_23378_additional_1.map.gz emd_23378_additional_1.map.gz emd_23378_additional_2.map.gz emd_23378_additional_2.map.gz | 435.1 MB 498.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23378 http://ftp.pdbj.org/pub/emdb/structures/EMD-23378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23378 | HTTPS FTP |
-Validation report
| Summary document |  emd_23378_validation.pdf.gz emd_23378_validation.pdf.gz | 428.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23378_full_validation.pdf.gz emd_23378_full_validation.pdf.gz | 428.4 KB | Display | |
| Data in XML |  emd_23378_validation.xml.gz emd_23378_validation.xml.gz | 9.9 KB | Display | |
| Data in CIF |  emd_23378_validation.cif.gz emd_23378_validation.cif.gz | 11.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23378 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23378 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23378 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23378 | HTTPS FTP |
-Related structure data
| Related structure data |  7lihMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23378.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23378.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of MAYV focused region | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.284 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
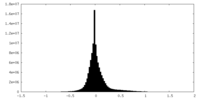
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: CryoEM density map of MAYV virion
| File | emd_23378_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of MAYV virion | ||||||||||||
| Projections & Slices |
| ||||||||||||
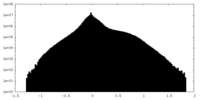
| Density Histograms |
-Additional map: CryoEM density map of E1-E2-E3-CP subunit.
| File | emd_23378_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of E1-E2-E3-CP subunit. | ||||||||||||
| Projections & Slices |
| ||||||||||||
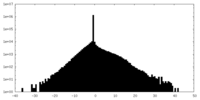
| Density Histograms |
- Sample components
Sample components
-Entire : Mayaro virus
| Entire | Name:  Mayaro virus Mayaro virus |
|---|---|
| Components |
|
-Supramolecule #1: Mayaro virus
| Supramolecule | Name: Mayaro virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 59301 / Sci species name: Mayaro virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Diameter: 700.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mayaro virus Mayaro virus |
| Molecular weight | Theoretical: 17.596111 KDa |
| Sequence | String: RERMCMKIEH DCIFEVKHEG KVTGYACLVG DKVMKPAHVP GVIDNADLAR LSYKKSSKYD LECAQIPVAM KSDASKYTHE KPEGHYNWH YGAVQYTGGR FTVPTGVGKP GDSGRPIFDN KGPVVAIVLG GANEGTRTAL SVVTWNKDMV TKITPEGTVE W UniProtKB: Structural polyprotein |
-Macromolecule #2: E1 protein
| Macromolecule | Name: E1 protein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mayaro virus Mayaro virus |
| Molecular weight | Theoretical: 47.182426 KDa |
| Sequence | String: YEHTAVIPNQ VGFPYKAHVA REGYSPLTLQ MQVVETSLEP TLNLEYITCD YKTKVPSPYV KCCGTAECRT QDKPEYKCAV FTGVYPFMW GGAYCFCDSE NTQMSEAYVE RADVCKHDYA AAYRAHTASL RAKIKVTYGT VNQTVEAYVN GDHAVTLAGT K FIFGPVST ...String: YEHTAVIPNQ VGFPYKAHVA REGYSPLTLQ MQVVETSLEP TLNLEYITCD YKTKVPSPYV KCCGTAECRT QDKPEYKCAV FTGVYPFMW GGAYCFCDSE NTQMSEAYVE RADVCKHDYA AAYRAHTASL RAKIKVTYGT VNQTVEAYVN GDHAVTLAGT K FIFGPVST AWTPFDTKIV VYKGEVYNQD FPPYGAGQPG RFGDIQSRTL DSKDLYANTG LKLARPAAGN IHVPYTQTPS GF KTWQKDR DSPLNAKAPF GCTIQTNPVR AMNCAVGNIP VSMDIADSAF TRLTDAPIIS ELLCTVSTCT HSSDFGGVAV LSY KVEKAG RCDVHSHSNV AVLQEVSIEA EGRSVIHFST ASAAPSFIVS VCSSRATCTA KCEPPKDHVV TYPANHNGIT LPDL SSTAM TWAQHLAGGV GLLIALAVLI LVIVTCITLR UniProtKB: Structural polyprotein |
-Macromolecule #3: E2 protein
| Macromolecule | Name: E2 protein / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mayaro virus Mayaro virus |
| Molecular weight | Theoretical: 45.54491 KDa |
| Sequence | String: NAYKLTRPYV AYCADCGMGH SCHSPAMIEN VQADATDGTL KIQFASQIGL TKTDTHDHTK IRYAEGHDIA EAARSTLKVH SSSECAVTG TMGHFILAKC PPGEVISVSF VDSKNEQRTC RIAYHHEQRL IGRERFTVRP HHGIELPCTT YQLTTAETSE E IDMHMPPD ...String: NAYKLTRPYV AYCADCGMGH SCHSPAMIEN VQADATDGTL KIQFASQIGL TKTDTHDHTK IRYAEGHDIA EAARSTLKVH SSSECAVTG TMGHFILAKC PPGEVISVSF VDSKNEQRTC RIAYHHEQRL IGRERFTVRP HHGIELPCTT YQLTTAETSE E IDMHMPPD IPDRTILSQQ SGNVKITVNG RTVKYSCSCG SKPSGTTTTD KTINSCTVDK CQAYVTSHTK WQFNSPFVPR AE QAERKGK VHIPFPLINT TCRVPLAPEA LVRSGKREAT LSLHPIHPTL LSYRTLGREP VFDEQWITTQ TEVTIPVPVE GVE YRWGNH KPQRLWSQLT TEGRAHGWPH EIIEYYYGLH PTTTIVVVIR VSVVVLLSFA ASVYMCVVAR TKCLTPYALT PGAV VPVTI GVLCC UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)