[English] 日本語
 Yorodumi
Yorodumi- EMDB-23372: The structure of the insect olfactory receptor OR5 from Machilis ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23372 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the insect olfactory receptor OR5 from Machilis hrabei. | |||||||||
 Map data Map data | Olfactory receptor OR5 from the insect species Machilis hrabei, apo state. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | olfactory receptor / ion channel / membrane protein | |||||||||
| Biological species |  Machilis hrabei (insect) Machilis hrabei (insect) | |||||||||
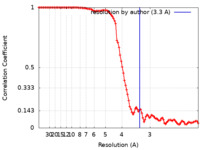
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | del Marmol J / Ruta V | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: The structural basis of odorant recognition in insect olfactory receptors. Authors: Josefina Del Mármol / Mackenzie A Yedlin / Vanessa Ruta /  Abstract: Olfactory systems must detect and discriminate amongst an enormous variety of odorants. To contend with this challenge, diverse species have converged on a common strategy in which odorant identity ...Olfactory systems must detect and discriminate amongst an enormous variety of odorants. To contend with this challenge, diverse species have converged on a common strategy in which odorant identity is encoded through the combinatorial activation of large families of olfactory receptors, thus allowing a finite number of receptors to detect a vast chemical world. Here we offer structural and mechanistic insight into how an individual olfactory receptor can flexibly recognize diverse odorants. We show that the olfactory receptor MhOR5 from the jumping bristletail Machilis hrabei assembles as a homotetrameric odorant-gated ion channel with broad chemical tuning. Using cryo-electron microscopy, we elucidated the structure of MhOR5 in multiple gating states, alone and in complex with two of its agonists-the odorant eugenol and the insect repellent DEET. Both ligands are recognized through distributed hydrophobic interactions within the same geometrically simple binding pocket located in the transmembrane region of each subunit, suggesting a structural logic for the promiscuous chemical sensitivity of this receptor. Mutation of individual residues lining the binding pocket predictably altered the sensitivity of MhOR5 to eugenol and DEET and broadly reconfigured the receptor's tuning. Together, our data support a model in which diverse odorants share the same structural determinants for binding, shedding light on the molecular recognition mechanisms that ultimately endow the olfactory system with its immense discriminatory capacity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23372.map.gz emd_23372.map.gz | 94.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23372-v30.xml emd-23372-v30.xml emd-23372.xml emd-23372.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23372_fsc.xml emd_23372_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_23372.png emd_23372.png | 101.7 KB | ||
| Filedesc metadata |  emd-23372.cif.gz emd-23372.cif.gz | 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23372 http://ftp.pdbj.org/pub/emdb/structures/EMD-23372 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23372 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23372 | HTTPS FTP |
-Validation report
| Summary document |  emd_23372_validation.pdf.gz emd_23372_validation.pdf.gz | 513.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23372_full_validation.pdf.gz emd_23372_full_validation.pdf.gz | 512.8 KB | Display | |
| Data in XML |  emd_23372_validation.xml.gz emd_23372_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_23372_validation.cif.gz emd_23372_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23372 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23372 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23372 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23372 | HTTPS FTP |
-Related structure data
| Related structure data |  7licMC  7lidC  7ligC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23372.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23372.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Olfactory receptor OR5 from the insect species Machilis hrabei, apo state. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Homotetrameric olfactory receptor MhOR5
| Entire | Name: Homotetrameric olfactory receptor MhOR5 |
|---|---|
| Components |
|
-Supramolecule #1: Homotetrameric olfactory receptor MhOR5
| Supramolecule | Name: Homotetrameric olfactory receptor MhOR5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Machilis hrabei (insect) Machilis hrabei (insect) |
-Macromolecule #1: MhOR5
| Macromolecule | Name: MhOR5 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Machilis hrabei (insect) Machilis hrabei (insect) |
| Molecular weight | Theoretical: 53.736695 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGRAKIDVD SVDHTDDYIH LRKWIKRIGI ILRISGHWPF RLPHEKRNQH KSKFRQVYSC LVITLGFITC SCYCIGLCLS ESIAQALNN ITVTSYFLQS CVCYVSFIIN SRKLETLFNY LFENEVVGCP RGYKMSSIKT TLFRCKFVAF SLGILSFFGW L MWTLLPLA ...String: GPGRAKIDVD SVDHTDDYIH LRKWIKRIGI ILRISGHWPF RLPHEKRNQH KSKFRQVYSC LVITLGFITC SCYCIGLCLS ESIAQALNN ITVTSYFLQS CVCYVSFIIN SRKLETLFNY LFENEVVGCP RGYKMSSIKT TLFRCKFVAF SLGILSFFGW L MWTLLPLA VLVVDSGATG GGNQTSLRFV EAWYPFDTTT SPMNEVIAIY EAVAMIFLIT APMSSDIMFC VLMIFIVEHL KC LGMAIEC TLKGISTNQH QNIGFDDSVS DVNVQRRIVI GKESPIQSIH VPIKECSRQS SDAVFREKRH GTHHQIIRSH NYT DATSLC NIVDSHVKIY RTMEIVQSVY SSYFATLFFT SCLAVCALAY FLAATSTSFT RVPGMVLYLM YIFLRIFLLC LLAT EVAEQ GLNLCHAGYS SKLVLASDHV RSTIQAIATR AQIPLSITGA RFFTVNLSFL ASMAGVMLTY FIVLLQVNAK PKP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.51 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)