+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23251 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a ferrichrome importer FhuCDB from E. coli | |||||||||
 Map data Map data | ferrichrome importer FhuCDB | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC importer / siderophore / cryo-EM / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type ferric hydroxamate transporter / ABC-type ferric hydroxamate transporter activity / ferric iron transmembrane transporter activity / ferric-hydroxamate import into cell / Metal ion assimilation from the host / iron ion import across plasma membrane / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing ...ABC-type ferric hydroxamate transporter / ABC-type ferric hydroxamate transporter activity / ferric iron transmembrane transporter activity / ferric-hydroxamate import into cell / Metal ion assimilation from the host / iron ion import across plasma membrane / siderophore-iron import into cell / siderophore uptake transmembrane transporter activity / transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cellular response to iron ion / outer membrane-bounded periplasmic space / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
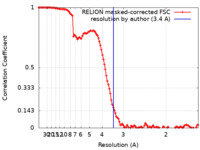
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Hu W / Zheng H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM reveals unique structural features of the FhuCDB Escherichia coli ferrichrome importer. Authors: Wenxin Hu / Hongjin Zheng /  Abstract: As one of the most elegant biological processes developed in bacteria, the siderophore-mediated iron uptake demands the action of specific ATP-binding cassette (ABC) importers. Although extensive ...As one of the most elegant biological processes developed in bacteria, the siderophore-mediated iron uptake demands the action of specific ATP-binding cassette (ABC) importers. Although extensive studies have been done on various ABC importers, the molecular basis of these iron-chelated-siderophore importers are still not fully understood. Here, we report the structure of a ferrichrome importer FhuCDB from Escherichia coli at 3.4 Å resolution determined by cryo electron microscopy. The structure revealed a monomeric membrane subunit of FhuB with a substrate translocation pathway in the middle. In the pathway, there were unique arrangements of residues, especially layers of methionines. Important residues found in the structure were interrogated by mutagenesis and functional studies. Surprisingly, the importer's ATPase activity was decreased upon FhuD binding, which deviated from the current understanding about bacterial ABC importers. In summary, to the best of our knowledge, these studies not only reveal a new structural twist in the type II ABC importer subfamily, but also provide biological insights in the transport of iron-chelated siderophores. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23251.map.gz emd_23251.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23251-v30.xml emd-23251-v30.xml emd-23251.xml emd-23251.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23251_fsc.xml emd_23251_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_23251.png emd_23251.png | 200.2 KB | ||
| Filedesc metadata |  emd-23251.cif.gz emd-23251.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23251 http://ftp.pdbj.org/pub/emdb/structures/EMD-23251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23251 | HTTPS FTP |
-Related structure data
| Related structure data |  7lb8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23251.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23251.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ferrichrome importer FhuCDB | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.799 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : holocomplex of ferrichrome importer FhuCDB
| Entire | Name: holocomplex of ferrichrome importer FhuCDB |
|---|---|
| Components |
|
-Supramolecule #1: holocomplex of ferrichrome importer FhuCDB
| Supramolecule | Name: holocomplex of ferrichrome importer FhuCDB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Iron(3+)-hydroxamate import system permease protein FhuB
| Macromolecule | Name: Iron(3+)-hydroxamate import system permease protein FhuB type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 71.574867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKRIALFPA LLLALLVIVA TALTWMNFSQ ALPRSQWAQA AWSPNINVIE QMIFHYSLLP RLAISLLVGA GLGLVGVLFQ QVLRNPLAE PTTLGVATGA QLGITVTTLW AIPGAMASQF AALAGACVVG LIVFGVAWGK RLSPVTLILA GLVVSLYCGA I NQLLVIFH ...String: MSKRIALFPA LLLALLVIVA TALTWMNFSQ ALPRSQWAQA AWSPNINVIE QMIFHYSLLP RLAISLLVGA GLGLVGVLFQ QVLRNPLAE PTTLGVATGA QLGITVTTLW AIPGAMASQF AALAGACVVG LIVFGVAWGK RLSPVTLILA GLVVSLYCGA I NQLLVIFH HDQLQSMFLW STGTLTQTDW GGVERLWPQL LGGVMLTLLL LRPLTLMGLD DGVARNLGLA LSLARLAALS LA IVISALL VNAVGIIGFI GLFAPLLAKM LGARRLLPRL MLASLIGALI LWLSDQIILW LTRVWMEVST GSVTALIGAP LLL WLLPRL RSISAPDMKV NDRVATERQH VLAFALAGGV LLLMAVVVAL SFGRDAHGWT WASGALLDDL MPWRWPRIMA ALFA GVMLA VAGCIIQRLT GNPMASPEVL GISSGAAFGV VLMLFLVPGN AFGWLLPAGS LGAAVTLLII MIAAGRGGFS PHRML LAGM ALSTAFTMLL MMLQASGDPR MAQVLTWISG STYNATDAQV WRTGIVMVIL LAITPLCRRW LTILPLGGDT ARAVGM ALT PTRIALLLLA ACLTATATMT IGPLSFVGLM APHIARMMGF RRTMPHIVIS ALVGGLLLVF ADWCGRMVLF PFQIPAG LL STFIGAPYFI YLLRKQSRHH HHHHHH UniProtKB: Iron(3+)-hydroxamate import system permease protein FhuB |
-Macromolecule #2: Iron(3+)-hydroxamate-binding protein FhuD
| Macromolecule | Name: Iron(3+)-hydroxamate-binding protein FhuD / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.030258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGLPLISRR RLLTAMALSP LLWQMNTAHA AAIDPNRIVA LEWLPVELLL ALGIVPYGVA DTINYRLWVS EPPLPDSVID VGLRTEPNL ELLTTMRPSF MVWSAGYGPS PEMLARIAPG RGFNFSDGKQ PLAMARKSLT EMADLLNLQS AAETHLAQYE D FIRSMKPR ...String: MSGLPLISRR RLLTAMALSP LLWQMNTAHA AAIDPNRIVA LEWLPVELLL ALGIVPYGVA DTINYRLWVS EPPLPDSVID VGLRTEPNL ELLTTMRPSF MVWSAGYGPS PEMLARIAPG RGFNFSDGKQ PLAMARKSLT EMADLLNLQS AAETHLAQYE D FIRSMKPR FVKRGARPLL LTTLIDPRHM LVFGPNSLFQ EILDEYGIPN AWQGETNFWG STAVSIDRLA AYKDVDVLCF DH DNSKDMD ALMATPLWQA MPFVRAGRFQ RVPAVWFYGA TLSAMHFVRV LDNAIGGKA UniProtKB: Iron(3+)-hydroxamate-binding protein FhuD |
-Macromolecule #3: Iron(3+)-hydroxamate import ATP-binding protein FhuC
| Macromolecule | Name: Iron(3+)-hydroxamate import ATP-binding protein FhuC / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type ferric hydroxamate transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.921424 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQEYTNHSDT TFALRNISFR VPGRTLLHPL SLTFPAGKVT GLIGHNGSGK STLLKMLGRH QPPSEGEILL DAQPLESWSS KAFARKVAY LPQQLPPAEG MTVRELVAIG RYPWHGALGR FGAADREKVE EAISLVGLKP LAHRLVDSLS GGERQRAWIA M LVAQDSRC ...String: MQEYTNHSDT TFALRNISFR VPGRTLLHPL SLTFPAGKVT GLIGHNGSGK STLLKMLGRH QPPSEGEILL DAQPLESWSS KAFARKVAY LPQQLPPAEG MTVRELVAIG RYPWHGALGR FGAADREKVE EAISLVGLKP LAHRLVDSLS GGERQRAWIA M LVAQDSRC LLLDEPTSAL DIAHQVDVLS LVHRLSQERG LTVIAVLHDI NMAARYCDYL VALRGGEMIA QGTPAEIMRG ET LEMIYGI PMGILPHPAG AAPVSFVY UniProtKB: Iron(3+)-hydroxamate import ATP-binding protein FhuC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)