[English] 日本語
 Yorodumi
Yorodumi- EMDB-23062: Cryo-EM structure of EfPiwiA(MID/PIWI domains)-piRNA-long-target ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23062 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of EfPiwiA(MID/PIWI domains)-piRNA-long-target complex | |||||||||
 Map data Map data | EfPiwi (MID/PIWI domains)-piRNA-long-target complex full map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulatory ncRNA-mediated gene silencing / cell differentiation / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Ephydatia fluviatilis (invertebrata) / synthetic construct (others) Ephydatia fluviatilis (invertebrata) / synthetic construct (others) | |||||||||
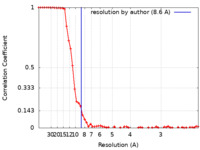
| Method | single particle reconstruction / cryo EM / Resolution: 8.6 Å | |||||||||
 Authors Authors | Chowdhury S / Anzelon TA / MacRae IJ / Lander GC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis for piRNA targeting. Authors: Todd A Anzelon / Saikat Chowdhury / Siobhan M Hughes / Yao Xiao / Gabriel C Lander / Ian J MacRae /   Abstract: PIWI proteins use PIWI-interacting RNAs (piRNAs) to identify and silence transposable elements and thereby maintain genome integrity between metazoan generations. The targeting of transposable ...PIWI proteins use PIWI-interacting RNAs (piRNAs) to identify and silence transposable elements and thereby maintain genome integrity between metazoan generations. The targeting of transposable elements by PIWI has been compared to mRNA target recognition by Argonaute proteins, which use microRNA (miRNA) guides, but the extent to which piRNAs resemble miRNAs is not known. Here we present cryo-electron microscopy structures of a PIWI-piRNA complex from the sponge Ephydatia fluviatilis with and without target RNAs, and a biochemical analysis of target recognition. Mirroring Argonaute, PIWI identifies targets using the piRNA seed region. However, PIWI creates a much weaker seed so that stable target association requires further piRNA-target pairing, making piRNAs less promiscuous than miRNAs. Beyond the seed, the structure of PIWI facilitates piRNA-target pairing in a manner that is tolerant of mismatches, leading to long-lived PIWI-piRNA-target interactions that may accumulate on transposable-element transcripts. PIWI ensures targeting fidelity by physically blocking the propagation of piRNA-target interactions in the absence of faithful seed pairing, and by requiring an extended piRNA-target duplex to reach an endonucleolytically active conformation. PIWI proteins thereby minimize off-targeting cellular mRNAs while defending against evolving genomic threats. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23062.map.gz emd_23062.map.gz | 14.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23062-v30.xml emd-23062-v30.xml emd-23062.xml emd-23062.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23062_fsc.xml emd_23062_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23062.png emd_23062.png | 33.9 KB | ||
| Masks |  emd_23062_msk_1.map emd_23062_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Others |  emd_23062_half_map_1.map.gz emd_23062_half_map_1.map.gz emd_23062_half_map_2.map.gz emd_23062_half_map_2.map.gz | 14.7 MB 14.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23062 http://ftp.pdbj.org/pub/emdb/structures/EMD-23062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23062 | HTTPS FTP |
-Validation report
| Summary document |  emd_23062_validation.pdf.gz emd_23062_validation.pdf.gz | 381.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23062_full_validation.pdf.gz emd_23062_full_validation.pdf.gz | 380.9 KB | Display | |
| Data in XML |  emd_23062_validation.xml.gz emd_23062_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_23062_validation.cif.gz emd_23062_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23062 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23062 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23062.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23062.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfPiwi (MID/PIWI domains)-piRNA-long-target complex full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
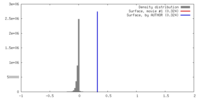
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23062_msk_1.map emd_23062_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
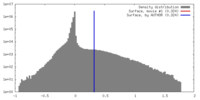
| Density Histograms |
-Half map: EfPiwi (MID/PIWI domains)-piRNA-long-target complex odd half map
| File | emd_23062_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfPiwi (MID/PIWI domains)-piRNA-long-target complex odd half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
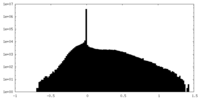
| Density Histograms |
-Half map: EfPiwi (MID/PIWI domains)-piRNA-long-target complex even half map
| File | emd_23062_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfPiwi (MID/PIWI domains)-piRNA-long-target complex even half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EfPiwiA(MID/PIWI domains)-piRNA-long-target complex
| Entire | Name: EfPiwiA(MID/PIWI domains)-piRNA-long-target complex |
|---|---|
| Components |
|
-Supramolecule #1: EfPiwiA(MID/PIWI domains)-piRNA-long-target complex
| Supramolecule | Name: EfPiwiA(MID/PIWI domains)-piRNA-long-target complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 45 KDa |
-Supramolecule #2: PiwiA(MID/PIWI domains)
| Supramolecule | Name: PiwiA(MID/PIWI domains) / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Ephydatia fluviatilis (invertebrata) Ephydatia fluviatilis (invertebrata) |
| Recombinant expression | Organism:  |
-Supramolecule #3: piRNA-long-target
| Supramolecule | Name: piRNA-long-target / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 97 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: Freezing was carried out in a cold room at 4 degree C and relative humidity between 95%-98%. 3.5 uL sample was applied to plasma cleaned grid and manually blotted with Whatman 1 filter paper ...Details: Freezing was carried out in a cold room at 4 degree C and relative humidity between 95%-98%. 3.5 uL sample was applied to plasma cleaned grid and manually blotted with Whatman 1 filter paper for 5-7 sec before plunge freezing in liquid ethane at -179 degree C.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 77.0 K / Max: 79.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-48 / Number real images: 1765 / Average exposure time: 12.0 sec. / Average electron dose: 47.33 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)