+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22931 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The capsid of Myoviridae Phage XM1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Myoviridae / Vibrio phage / Capsid / HK97 fold / VIRUS | |||||||||
| Biological species |  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wang Z / Klose T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structure of Vibrio phage XM1, a simple contractile DNA injection machine Authors: Wang Z / Fokine A / Guo X / Jiang W / Rossmann MG / Kuhn RJ / Luo ZH / Klose T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22931.map.gz emd_22931.map.gz | 1.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22931-v30.xml emd-22931-v30.xml emd-22931.xml emd-22931.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
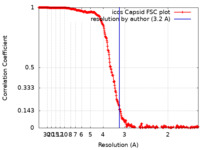
| FSC (resolution estimation) |  emd_22931_fsc.xml emd_22931_fsc.xml | 33.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_22931.png emd_22931.png | 166.1 KB | ||
| Filedesc metadata |  emd-22931.cif.gz emd-22931.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22931 http://ftp.pdbj.org/pub/emdb/structures/EMD-22931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22931 | HTTPS FTP |
-Related structure data
| Related structure data |  7kmxMC  7kh1C  7kjkC  7klnC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22931.map.gz / Format: CCP4 / Size: 3.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22931.map.gz / Format: CCP4 / Size: 3.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Vibrio phage XM1
| Entire | Name:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Vibrio phage XM1
| Supramolecule | Name: Vibrio phage XM1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2748688 / Sci species name: Vibrio phage XM1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Vibrio rotiferianus (bacteria) Vibrio rotiferianus (bacteria) |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 640.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Minor capsid protein
| Macromolecule | Name: Minor capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 16.823863 KDa |
| Sequence | String: MAFNNAVLQE VSDLPAGEVI KASPHNVSAF EVFQNGLIEG RFVKFDAGSI DILDASATPT IAGIAKRKVT GEIGPGVYST SGIEIDQVA EVINFGFATV TVQDAAAPSK YDPVYAINLD SAEAGKATEN SGATGALAVA DCVFWEQKAA NVWLVRMNKF L |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 35.512773 KDa |
| Sequence | String: MKKDLKFIKS VYDLKSFSDK AAFAKKTFKD EGGIILARNL EHVSSEIFTQ EFAGLTFLQG GIVVNNEGGY ATSVTKLKLK AEGGFRESG NDTNTTGKIT LSGESDSIPV FTLEGESDWS EIELKQAELQ NVNLPSRYFE AHAELYNRKI DELGYLGQTR T DGTQKTLG ...String: MKKDLKFIKS VYDLKSFSDK AAFAKKTFKD EGGIILARNL EHVSSEIFTQ EFAGLTFLQG GIVVNNEGGY ATSVTKLKLK AEGGFRESG NDTNTTGKIT LSGESDSIPV FTLEGESDWS EIELKQAELQ NVNLPSRYFE AHAELYNRKI DELGYLGQTR T DGTQKTLG LLNYGFVASG AGDTAANLSG DNLYQAIADL ITDQWAGVFN VETYKADRVV MPDTVYNICA KKILNSNGSE MS VLRALMT NFPTVTFGLT TKARDVGGTS RTTAYSSNRR AMQMRIPTPL NVSSVDQRGF KYYVESYFGV AGLDVIEDTA GRH LTGL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM Tris, pH 7.5, 100 mM NaCl, 8 mM MgSO4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-16 (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7kmx: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)