[English] 日本語
 Yorodumi
Yorodumi- EMDB-21670: Large ribosomal subunit of Halococcus morrhuae, an archaeon with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21670 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Large ribosomal subunit of Halococcus morrhuae, an archaeon with an unusual 5S rRNA insertion | |||||||||||||||
 Map data Map data | Final map of Halococcus morrhuae large subunit | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Halococcus morrhuae (archaea) Halococcus morrhuae (archaea) | |||||||||||||||
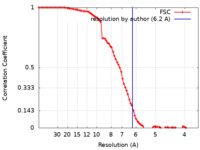
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||||||||
 Authors Authors | Tirumalai MR / Kaelber JT / Fox GE | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: FEBS Open Bio / Year: 2020 Journal: FEBS Open Bio / Year: 2020Title: Cryo-electron microscopy visualization of a large insertion in the 5S ribosomal RNA of the extremely halophilic archaeon Halococcus morrhuae. Authors: Madhan R Tirumalai / Jason T Kaelber / Donghyun R Park / Quyen Tran / George E Fox /  Abstract: The extreme halophile Halococcus morrhuae (ATCC 17082) contains a 108-nucleotide insertion in its 5S rRNA. Large rRNA expansions in Archaea are rare. This one almost doubles the length of the 5S rRNA. ...The extreme halophile Halococcus morrhuae (ATCC 17082) contains a 108-nucleotide insertion in its 5S rRNA. Large rRNA expansions in Archaea are rare. This one almost doubles the length of the 5S rRNA. In order to understand how such an insertion is accommodated in the ribosome, we obtained a cryo-electron microscopy reconstruction of the native large subunit at subnanometer resolution. The insertion site forms a four-way junction that fully preserves the canonical 5S rRNA structure. Moving away from the junction site, the inserted region is conformationally flexible and does not pack tightly against the large subunit. The high-salt requirement of the H. morrhuae ribosomes for their stability conflicted with the low-salt threshold for cryo-electron microscopy procedures. Despite this obstacle, this is the first cryo-electron microscopy map of Halococcus ribosomes. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21670.map.gz emd_21670.map.gz | 26.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21670-v30.xml emd-21670-v30.xml emd-21670.xml emd-21670.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21670_fsc.xml emd_21670_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_21670.png emd_21670.png | 104.1 KB | ||
| Masks |  emd_21670_msk_1.map emd_21670_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Others |  emd_21670_half_map_1.map.gz emd_21670_half_map_1.map.gz emd_21670_half_map_2.map.gz emd_21670_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21670 http://ftp.pdbj.org/pub/emdb/structures/EMD-21670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21670 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21670.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21670.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map of Halococcus morrhuae large subunit | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.74 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
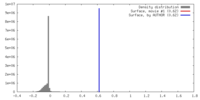
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21670_msk_1.map emd_21670_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
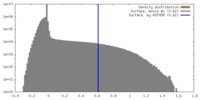
| Density Histograms |
-Half map: Unmasked unfiltered half-map A
| File | emd_21670_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked unfiltered half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
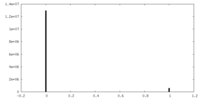
| Density Histograms |
-Half map: Unmasked unfiltered half-map B
| File | emd_21670_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked unfiltered half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
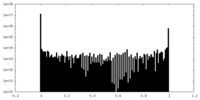
| Density Histograms |
- Sample components
Sample components
-Entire : Large ribosomal subunit of Halococcus morrhuae
| Entire | Name: Large ribosomal subunit of Halococcus morrhuae |
|---|---|
| Components |
|
-Supramolecule #1: Large ribosomal subunit of Halococcus morrhuae
| Supramolecule | Name: Large ribosomal subunit of Halococcus morrhuae / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Halococcus morrhuae (archaea) / Strain: ATCC 17082 Halococcus morrhuae (archaea) / Strain: ATCC 17082 |
| Molecular weight | Theoretical: 1.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
Details: pure water was added to the sample on the grid | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 2 microliters of sample were applied to the grid, then 6 microliters of pure water was applied to the reverse face of the grid.. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 902 / Average exposure time: 8.0 sec. / Average electron dose: 26.0 e/Å2 / Details: some images collected manually (no serialEM) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 5.716 µm / Calibrated defocus min: 0.915 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal magnification: 20000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)