[English] 日本語
 Yorodumi
Yorodumi- EMDB-2129: Cryo-electron tomography averaged map of microtubule doublet 9 in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomography averaged map of microtubule doublet 9 in the proximal region of Chlamydomonas axoneme | |||||||||
 Map data Map data | Reconstruction of outer doublet 9 of the Chlamydomonas axoneme in the proximal region. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | axoneme / dynein / Chlamydomonas / microtubule doublet | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 42.9 Å | |||||||||
 Authors Authors | Bui KH / Yagi T / Yamamoto R / Kamiya R / Ishikawa T | |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2012 Journal: J Cell Biol / Year: 2012Title: Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. Authors: Khanh Huy Bui / Toshiki Yagi / Ryosuke Yamamoto / Ritsu Kamiya / Takashi Ishikawa /  Abstract: Understanding the molecular architecture of the flagellum is crucial to elucidate the bending mechanism produced by this complex organelle. The current known structure of the flagellum has not yet ...Understanding the molecular architecture of the flagellum is crucial to elucidate the bending mechanism produced by this complex organelle. The current known structure of the flagellum has not yet been fully correlated with the complex composition and localization of flagellar components. Using cryoelectron tomography and subtomogram averaging while distinguishing each one of the nine outer doublet microtubules, we systematically collected and reconstructed the three-dimensional structures in different regions of the Chlamydomonas flagellum. We visualized the radial and longitudinal differences in the flagellum. One doublet showed a distinct structure, whereas the other eight were similar but not identical to each other. In the proximal region, some dyneins were missing or replaced by minor dyneins, and outer-inner arm dynein links were variable among different microtubule doublets. These findings shed light on the intricate organization of Chlamydomonas flagella, provide clues to the mechanism that produces asymmetric flagellar beating, and pose a new challenge for the functional study of the flagella. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2129.map.gz emd_2129.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2129-v30.xml emd-2129-v30.xml emd-2129.xml emd-2129.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2129.png emd_2129.png | 196.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2129 http://ftp.pdbj.org/pub/emdb/structures/EMD-2129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2129 | HTTPS FTP |
-Validation report
| Summary document |  emd_2129_validation.pdf.gz emd_2129_validation.pdf.gz | 220.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2129_full_validation.pdf.gz emd_2129_full_validation.pdf.gz | 219.9 KB | Display | |
| Data in XML |  emd_2129_validation.xml.gz emd_2129_validation.xml.gz | 6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2129 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2129 | HTTPS FTP |
-Related structure data
| Related structure data |  2113C  2114C  2115C  2116C  2117C  2118C  2119C  2120C  2121C  2122C  2123C  2124C  2125C  2126C  2127C  2128C  2130C  2131C  2132C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2129.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2129.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of outer doublet 9 of the Chlamydomonas axoneme in the proximal region. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
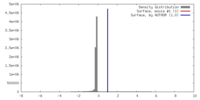
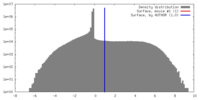
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Outer doublet 9 of Chlamydomonas axoneme in the proximal region
| Entire | Name: Outer doublet 9 of Chlamydomonas axoneme in the proximal region |
|---|---|
| Components |
|
-Supramolecule #1000: Outer doublet 9 of Chlamydomonas axoneme in the proximal region
| Supramolecule | Name: Outer doublet 9 of Chlamydomonas axoneme in the proximal region type: sample / ID: 1000 / Details: Flagella were isolated from Chlamydomonas. / Number unique components: 1 |
|---|
-Supramolecule #1: flagellum
| Supramolecule | Name: flagellum / type: organelle_or_cellular_component / ID: 1 / Name.synonym: axoneme,cilia / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 30 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 25 mM KCl, and 0.5% (wt/vol) polyethylene glycol (MW 20,000) |
| Grid | Details: 300 mesh Quantifoil Holey Carbon copper grid R2/1 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 110 K / Instrument: FEI VITROBOT MARK II / Method: Offset -3, blot 3 seconds, drain time 0 second |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Min: 93 K / Max: 118 K / Average: 98 K |
| Specialist optics | Energy filter - Name: Gatan Tridem / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Nov 26, 2010 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Number real images: 499 / Average electron dose: 60 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 19303 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder: Gatan 626, liquid nitrogen cooled / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
- Image processing
Image processing
| Details | R-weighted back projection using IMOD with fiducial markers. Average number of tilts used in the 3D reconstructions: 61. Average tomographic tilt angle increment: 2. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 42.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMOD, BSOFT, SPIDER, TOM, package, Matlab |
 Movie
Movie Controller
Controller


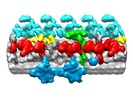
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)