[English] 日本語
 Yorodumi
Yorodumi- EMDB-21260: icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21260 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
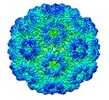
| Title | icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Brome mosaic virus (RNA 3 and 4) / VIRUS | |||||||||
| Function / homology | Bromovirus coat protein / Bromovirus coat protein / T=3 icosahedral viral capsid / host cell endoplasmic reticulum / viral nucleocapsid / ribonucleoprotein complex / structural molecule activity / RNA binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Brome mosaic virus Brome mosaic virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Beren C / Cui YX | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Genome organization and interaction with capsid protein in a multipartite RNA virus. Authors: Christian Beren / Yanxiang Cui / Antara Chakravarty / Xue Yang / A L N Rao / Charles M Knobler / Z Hong Zhou / William M Gelbart /  Abstract: We report the asymmetric reconstruction of the single-stranded RNA (ssRNA) content in one of the three otherwise identical virions of a multipartite RNA virus, brome mosaic virus (BMV). We exploit a ...We report the asymmetric reconstruction of the single-stranded RNA (ssRNA) content in one of the three otherwise identical virions of a multipartite RNA virus, brome mosaic virus (BMV). We exploit a sample consisting exclusively of particles with the same RNA content-specifically, RNAs 3 and 4-assembled in planta by agrobacterium-mediated transient expression. We find that the interior of the particle is nearly empty, with most of the RNA genome situated at the capsid shell. However, this density is disordered in the sense that the RNA is not associated with any particular structure but rather, with an ensemble of secondary/tertiary structures that interact with the capsid protein. Our results illustrate a fundamental difference between the ssRNA organization in the multipartite BMV viral capsid and the monopartite bacteriophages MS2 and Qβ for which a dominant RNA conformation is found inside the assembled viral capsids, with RNA density conserved even at the center of the particle. This can be understood in the context of the differing demands on their respective lifecycles: BMV must package separately each of several different RNA molecules and has been shown to replicate and package them in isolated, membrane-bound, cytoplasmic complexes, whereas the bacteriophages exploit sequence-specific "packaging signals" throughout the viral RNA to package their monopartite genomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21260.map.gz emd_21260.map.gz | 150.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21260-v30.xml emd-21260-v30.xml emd-21260.xml emd-21260.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21260.png emd_21260.png | 269.1 KB | ||
| Filedesc metadata |  emd-21260.cif.gz emd-21260.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21260 http://ftp.pdbj.org/pub/emdb/structures/EMD-21260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21260 | HTTPS FTP |
-Validation report
| Summary document |  emd_21260_validation.pdf.gz emd_21260_validation.pdf.gz | 594.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21260_full_validation.pdf.gz emd_21260_full_validation.pdf.gz | 594.2 KB | Display | |
| Data in XML |  emd_21260_validation.xml.gz emd_21260_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_21260_validation.cif.gz emd_21260_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21260 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21260 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21260 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21260 | HTTPS FTP |
-Related structure data
| Related structure data |  6vocMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21260.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21260.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Brome mosaic virus
| Entire | Name:   Brome mosaic virus Brome mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Brome mosaic virus
| Supramolecule | Name: Brome mosaic virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12302 / Sci species name: Brome mosaic virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Brome mosaic virus Brome mosaic virus |
| Molecular weight | Theoretical: 20.411525 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTSGTGKMT RAQRRAAARR NRWTARVQPV IVEPLAAGQG KAIKAIAGYS ISKWEASSDA ITAKATNAMS ITLPHELSSE KNKELKVGR VLLWLGLLPS VAGRIKACVA EKQAQAEAAF QVALAVADSS KEVVAAMYTD AFRGATLGDL LNLQIYLYAS E AVPAKAVV VHLEVEHVRP TFDDFFTPVY R UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Chromatic aberration corrector: none / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-30 / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 30.0 µm / Calibrated defocus min: 8.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















