[English] 日本語
 Yorodumi
Yorodumi- EMDB-21179: EM-Based Polyclonal Epitope Mapping. ConM-SOSIP Complexed with Pu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21179 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM-Based Polyclonal Epitope Mapping. ConM-SOSIP Complexed with Purified Polyclonal Fab (Grp 1, ID r2381 Wk22) | |||||||||
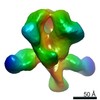
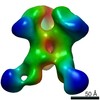
 Map data Map data | EM-based polyclonal epitope mapping, r2381 (Wk22-Fab) ConM-SOSIPv9; Negative stain EM map | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 17.89 Å | |||||||||
 Authors Authors | Ward AB / Antanasijevic A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2020 Journal: PLoS Pathog / Year: 2020Title: Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. Authors: Aleksandar Antanasijevic / George Ueda / Philip J M Brouwer / Jeffrey Copps / Deli Huang / Joel D Allen / Christopher A Cottrell / Anila Yasmeen / Leigh M Sewall / Ilja Bontjer / Thomas J ...Authors: Aleksandar Antanasijevic / George Ueda / Philip J M Brouwer / Jeffrey Copps / Deli Huang / Joel D Allen / Christopher A Cottrell / Anila Yasmeen / Leigh M Sewall / Ilja Bontjer / Thomas J Ketas / Hannah L Turner / Zachary T Berndsen / David C Montefiori / Per Johan Klasse / Max Crispin / David Nemazee / John P Moore / Rogier W Sanders / Neil P King / David Baker / Andrew B Ward /    Abstract: Two-component, self-assembling nanoparticles represent a versatile platform for multivalent presentation of viral antigens. Computational design of protein nanoparticles with differing sizes and ...Two-component, self-assembling nanoparticles represent a versatile platform for multivalent presentation of viral antigens. Computational design of protein nanoparticles with differing sizes and geometries enables combination with antigens of choice to test novel multimerization concepts in immunization strategies where the goal is to improve the induction and maturation of neutralizing antibody lineages. Here, we describe detailed antigenic, structural, and functional characterization of computationally designed tetrahedral, octahedral, and icosahedral nanoparticle immunogens displaying trimeric HIV envelope glycoprotein (Env) ectodomains. Env trimers, based on subtype A (BG505) or consensus group M (ConM) sequences and engineered with SOSIP stabilizing mutations, were fused to an underlying trimeric building block of each nanoparticle. Initial screening yielded one icosahedral and two tetrahedral nanoparticle candidates, capable of presenting twenty or four copies of the Env trimer. A number of analyses, including detailed structural characterization by cryo-EM, demonstrated that the nanoparticle immunogens possessed the intended structural and antigenic properties. When the immunogenicity of ConM-SOSIP trimers presented on a two-component tetrahedral nanoparticle or as soluble proteins were compared in rabbits, the two immunogens elicited similar serum antibody binding titers against the trimer component. Neutralizing antibody titers were slightly elevated in the animals given the nanoparticle immunogen and were initially more focused to the trimer apex. Altogether, our findings indicate that tetrahedral nanoparticles can be successfully applied for presentation of HIV Env trimer immunogens; however, the optimal implementation to different immunization strategies remains to be determined. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21179.map.gz emd_21179.map.gz | 20.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21179-v30.xml emd-21179-v30.xml emd-21179.xml emd-21179.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21179.png emd_21179.png | 47.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21179 http://ftp.pdbj.org/pub/emdb/structures/EMD-21179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21179 | HTTPS FTP |
-Validation report
| Summary document |  emd_21179_validation.pdf.gz emd_21179_validation.pdf.gz | 78 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21179_full_validation.pdf.gz emd_21179_full_validation.pdf.gz | 77.1 KB | Display | |
| Data in XML |  emd_21179_validation.xml.gz emd_21179_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21179 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21179 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21179.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21179.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM-based polyclonal epitope mapping, r2381 (Wk22-Fab) ConM-SOSIPv9; Negative stain EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ConM-SOSIP.v9 complexed with purified polyclonal Fab from an immu...
| Entire | Name: ConM-SOSIP.v9 complexed with purified polyclonal Fab from an immunized rabbit (Grp 1, ID r2381, Wk22) |
|---|---|
| Components |
|
-Supramolecule #1: ConM-SOSIP.v9 complexed with purified polyclonal Fab from an immu...
| Supramolecule | Name: ConM-SOSIP.v9 complexed with purified polyclonal Fab from an immunized rabbit (Grp 1, ID r2381, Wk22) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: ConM-SOSIP.v9
| Macromolecule | Name: ConM-SOSIP.v9 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDACTTLFCA SDAKAYDTEK RNVWATHCCV PTDPNPQEIV LENVTENFNM WKNNMVEQMH EDIISLWDQS LKPCVKLTPL CVTLNCTDVN ATNNTTNNEE IKNCSFNITT ELRDKKKKVY ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDACTTLFCA SDAKAYDTEK RNVWATHCCV PTDPNPQEIV LENVTENFNM WKNNMVEQMH EDIISLWDQS LKPCVKLTPL CVTLNCTDVN ATNNTTNNEE IKNCSFNITT ELRDKKKKVY ALFYKLDVVP IDDNNSYRLI NCNTSAITQA CPKVSFEPIP IHYCAPAGFA ILKCNDKKFN GTGPCKNVST VQCTHGIKPV VSTQLLLNGS LAEEEIIIRS ENITNNAKTI IVQLNESVEI NCTRPNNNTV KSIRIGPGQW FYYTGDIIGD IRQAHCNISR TKWNKTLQQV AKKLREHFNK TIIFNQSSGG DLEITTHSFN CGGEFFYCNT SELFNSTWNG TNNTITLPCR IKQIINMWQR VGQAMYAPPI EGKIRCTSNI TGLLLTRDGG NNNTETFRPG GGDMRDNWRS ELYKYKVVKI EPLGVAPTRC KRRVVERRRR RRAVGIGAVS LGFLGAAGST MGAASMTLTV QARNLLSGIV QQQSNLLCAP ECQQHLLQDT HWGIKQLQAR VLAVEHYLKD QQLLGIWGCS GKLICCTNVP WNSSWSNKSQ DEIWDNMTWM EWDKEINNYT DIIYSLIEES QNQQEKNEQE LLALD |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS buffer, pH 7.4 | |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Sample diluted to 0.05 mg/mL. 3 uL was applied onto the grid, blotted off, and then stained with 2% uranyl formate for 60 seconds. | |||||||||
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Details | ConM-SOSIP.v9 was complexed with the purified polyclonal Fab overnight. The complex was purified using SEC and concentrated. The complex was then diluted in TBS to 0.05mg/ml to be loaded onto an imaging grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera
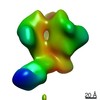












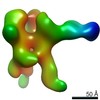
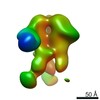
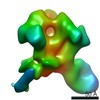


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















