+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-2100 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | |||||||||
 マップデータ マップデータ | reconstruction of rotavirus DLP+VP1 (polymerase) | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | rotavirus / dsRNA-dependent / polymerase | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報viral genome replication / virion component / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / RNA binding 類似検索 - 分子機能 | |||||||||
| 生物種 |  Bovine rotavirus (ウイルス) Bovine rotavirus (ウイルス) | |||||||||
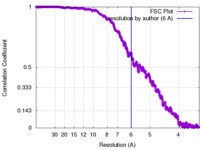
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.0 Å | |||||||||
 データ登録者 データ登録者 | Estrozi LF / Settembre EC / Goret G / McClain B / Zhang X / Chen JZ / Grigorieff N / Harrison SC | |||||||||
 引用 引用 |  ジャーナル: J Mol Biol / 年: 2013 ジャーナル: J Mol Biol / 年: 2013タイトル: Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. 著者: Leandro F Estrozi / Ethan C Settembre / Gaël Goret / Brian McClain / Xing Zhang / James Z Chen / Nikolaus Grigorieff / Stephen C Harrison /  要旨: Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense mRNA emerges into the intracellular milieu. During infectious entry of a ...Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense mRNA emerges into the intracellular milieu. During infectious entry of a rotavirus particle, the outer layer of its three-layer structure dissociates, delivering the inner double-layered particle (DLP) into the cytosol. DLP structures determined by X-ray crystallography and electron cryomicroscopy (cryoEM) show that the RNA coils uniformly into the particle interior, avoiding a "fivefold hub" of more structured density projecting inward from the VP2 shell of the DLP along each of the twelve 5-fold axes. Analysis of the X-ray crystallographic electron density map suggested that principal contributors to the hub are the N-terminal arms of VP2, but reexamination of the cryoEM map has shown that many features come from a molecule of VP1, randomly occupying five equivalent and partly overlapping positions. We confirm here that the electron density in the X-ray map leads to the same conclusion, and we describe the functional implications of the orientation and position of the polymerase. The exit channel for the nascent transcript directs the nascent transcript toward an opening along the 5-fold axis. The template strand enters from within the particle, and the dsRNA product of the initial replication step exits in a direction tangential to the inner surface of the VP2 shell, allowing it to coil optimally within the DLP. The polymerases of reoviruses appear to have similar positions and functional orientations. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_2100.map.gz emd_2100.map.gz | 7.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-2100-v30.xml emd-2100-v30.xml emd-2100.xml emd-2100.xml | 13.4 KB 13.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_2100_fsc.xml emd_2100_fsc.xml | 130.3 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_2100.png emd_2100.png | 207 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2100 http://ftp.pdbj.org/pub/emdb/structures/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2100 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_2100_validation.pdf.gz emd_2100_validation.pdf.gz | 259.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_2100_full_validation.pdf.gz emd_2100_full_validation.pdf.gz | 258.8 KB | 表示 | |
| XML形式データ |  emd_2100_validation.xml.gz emd_2100_validation.xml.gz | 47.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2100 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2100 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_2100.map.gz / 形式: CCP4 / 大きさ: 20.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_2100.map.gz / 形式: CCP4 / 大きさ: 20.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | reconstruction of rotavirus DLP+VP1 (polymerase) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 これらの図は立方格子座標系で作成されたものです | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.69643 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Rotavirus DLP+VP1
| 全体 | 名称: Rotavirus DLP+VP1 |
|---|---|
| 要素 |
|
-超分子 #1000: Rotavirus DLP+VP1
| 超分子 | 名称: Rotavirus DLP+VP1 / タイプ: sample / ID: 1000 集合状態: 780 molecules of VP6 form a DLP particle with 12 molecules of VP1, 120 molecules of VP2, 12 molecules of VP3 and 11 dsRNA molecules Number unique components: 5 |
|---|
-分子 #1: Rotavirus polymerase (VP1)
| 分子 | 名称: Rotavirus polymerase (VP1) / タイプ: protein_or_peptide / ID: 1 / Name.synonym: VP1 詳細: The icosahedral 3D reconstruction of rotavirus DLP shows extra-density near the 5-fold axis corresponding to one copy of VP1 attached to the DLP inner surface. コピー数: 11 / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  Bovine rotavirus (ウイルス) / 別称: Rotavirus Bovine rotavirus (ウイルス) / 別称: Rotavirus |
| 分子量 | 理論値: 126.326 KDa |
-分子 #2: VP1
| 分子 | 名称: VP1 / タイプ: protein_or_peptide / ID: 2 / Name.synonym: VP1 / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  Bovine rotavirus (ウイルス) / 別称: Rotavirus Bovine rotavirus (ウイルス) / 別称: Rotavirus |
-分子 #3: VP2
| 分子 | 名称: VP2 / タイプ: protein_or_peptide / ID: 3 / Name.synonym: VP2 / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  Bovine rotavirus (ウイルス) / 別称: Rotavirus Bovine rotavirus (ウイルス) / 別称: Rotavirus |
-分子 #4: VP3
| 分子 | 名称: VP3 / タイプ: protein_or_peptide / ID: 4 / Name.synonym: VP3 / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  Bovine rotavirus (ウイルス) / 別称: Rotavirus Bovine rotavirus (ウイルス) / 別称: Rotavirus |
-分子 #5: dsRNA
| 分子 | 名称: dsRNA / タイプ: rna / ID: 5 / Name.synonym: dsRNA / 分類: OTHER / Structure: OTHER / Synthetic?: No |
|---|---|
| 由来(天然) | 生物種:  Bovine rotavirus (ウイルス) / 別称: Rotavirus Bovine rotavirus (ウイルス) / 別称: Rotavirus |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 5 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | 詳細: Lacy carbon and C-flat |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 30 % / 装置: HOMEMADE PLUNGER 詳細: Vitrification instrument: Home-made. Vitrification carried out in air at room temperature 手法: Blot for 3 seconds before plunging |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI F30 |
|---|---|
| 温度 | 平均: 90 K |
| アライメント法 | Legacy - 非点収差: Objective lens astigmatism was corrected |
| 日付 | 2007年6月1日 |
| 撮影 | カテゴリ: FILM / フィルム・検出器のモデル: KODAK SO-163 FILM / デジタル化 - スキャナー: ZEISS SCAI / デジタル化 - サンプリング間隔: 7 µm / 実像数: 386 / 平均電子線量: 15 e/Å2 / Od range: 1 / ビット/ピクセル: 8 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 倍率(補正後): 56540 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2 mm / 最大 デフォーカス(公称値): 3.5 µm / 最小 デフォーカス(公称値): 1.1 µm / 倍率(公称値): 59000 |
| 試料ステージ | 試料ホルダー: Eucentric, side-entry / 試料ホルダーモデル: GATAN LIQUID NITROGEN |
| 実験機器 |  モデル: Tecnai F30 / 画像提供: FEI Company |
- 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Chain ID: A |
|---|---|
| ソフトウェア | 名称: URO and VEDA |
| 詳細 | Protocol: Rigid body. The Fourier coefficients of VP1 were downweighted by a factor 5 in order to prevent VP1 from being "attracted" by the stronger density of the VP2/6 layer during the fit. matrix 1 0.286272 -0.958148 -0.000156 150.76188 0.573968 0.171358 0.800748 -71.02608 -0.767209 -0.229322 0.599001 -168.57402 matrix 2 -0.084128 -0.546853 -0.832991 184.93534 0.025364 -0.836859 0.546831 15.73129 -0.996132 0.024876 0.084274 -147.45338 matrix 3 0.245240 0.395934 -0.884926 125.30383 -0.558295 -0.688567 -0.462799 80.75011 -0.792568 0.607546 0.052184 -184.30794 matrix 4 0.819204 0.567290 -0.084182 54.27816 -0.370407 0.411304 -0.832843 34.17286 -0.437839 0.713450 0.547070 -228.20418 matrix 5 0.844563 -0.269594 0.462637 70.01368 0.329383 0.942768 -0.051920 -59.62970 -0.422162 0.196234 0.885026 -218.47982 |
| 精密化 | 空間: RECIPROCAL / プロトコル: RIGID BODY FIT / 当てはまり具合の基準: Correlation coefficient |
| 得られたモデル |  PDB-4au6: |
 ムービー
ムービー コントローラー
コントローラー










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)