[English] 日本語
 Yorodumi
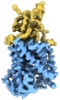
Yorodumi- EMDB-20807: Cryo-EM structure of the potassium-chloride cotransporter KCC4 in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20807 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs | |||||||||
 Map data Map data | Potassium-chloride cotransporter KCC4 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Potassium chloride cotransporter / SLC12 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCation-coupled Chloride cotransporters / chloride:monoatomic cation symporter activity / potassium:chloride symporter activity / potassium ion import across plasma membrane / potassium ion transmembrane transport / chloride transmembrane transport / cellular response to glucose stimulus / protein kinase binding / protein-containing complex / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Reid MS / Kern DM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs. Authors: Michelle S Reid / David M Kern / Stephen Graf Brohawn /  Abstract: Cation-chloride-cotransporters (CCCs) catalyze transport of Cl with K and/or Naacross cellular membranes. CCCs play roles in cellular volume regulation, neural development and function, audition, ...Cation-chloride-cotransporters (CCCs) catalyze transport of Cl with K and/or Naacross cellular membranes. CCCs play roles in cellular volume regulation, neural development and function, audition, regulation of blood pressure, and renal function. CCCs are targets of clinically important drugs including loop diuretics and their disruption has been implicated in pathophysiology including epilepsy, hearing loss, and the genetic disorders Andermann, Gitelman, and Bartter syndromes. Here we present the structure of a CCC, the K-Cl cotransporter (KCC) KCC4, in lipid nanodiscs determined by cryo-EM. The structure, captured in an inside-open conformation, reveals the architecture of KCCs including an extracellular domain poised to regulate transport activity through an outer gate. We identify binding sites for substrate K and Cl ions, demonstrate the importance of key coordinating residues for transporter activity, and provide a structural explanation for varied substrate specificity and ion transport ratio among CCCs. These results provide mechanistic insight into the function and regulation of a physiologically important transporter family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20807.map.gz emd_20807.map.gz | 20.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20807-v30.xml emd-20807-v30.xml emd-20807.xml emd-20807.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20807.png emd_20807.png | 119.9 KB | ||
| Filedesc metadata |  emd-20807.cif.gz emd-20807.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20807 http://ftp.pdbj.org/pub/emdb/structures/EMD-20807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20807 | HTTPS FTP |
-Validation report
| Summary document |  emd_20807_validation.pdf.gz emd_20807_validation.pdf.gz | 447.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20807_full_validation.pdf.gz emd_20807_full_validation.pdf.gz | 447.2 KB | Display | |
| Data in XML |  emd_20807_validation.xml.gz emd_20807_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_20807_validation.cif.gz emd_20807_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20807 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20807 | HTTPS FTP |
-Related structure data
| Related structure data |  6uknMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10394 (Title: Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs EMPIAR-10394 (Title: Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscsData size: 1.1 TB Data #1: Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20807.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20807.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Potassium-chloride cotransporter KCC4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.137 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : KCC4 in lipid nanodiscs
| Entire | Name: KCC4 in lipid nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: KCC4 in lipid nanodiscs
| Supramolecule | Name: KCC4 in lipid nanodiscs / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 12 member 7
| Macromolecule | Name: Solute carrier family 12 member 7 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 120.610961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPTNFTVVPV EARADGAGDE AAERTEEPES PESVDQTSPT PGDGNPRENS PFINNVEVER ESYFEGKNMA LFEEEMDSNP MVSSLLNKL ANYTNLSQGV VEHEEDEDSR RREVKAPRMG TFIGVYLPCL QNILGVILFL RLTWIVGAAG VMESFLIVAM C CTCTMLTA ...String: MPTNFTVVPV EARADGAGDE AAERTEEPES PESVDQTSPT PGDGNPRENS PFINNVEVER ESYFEGKNMA LFEEEMDSNP MVSSLLNKL ANYTNLSQGV VEHEEDEDSR RREVKAPRMG TFIGVYLPCL QNILGVILFL RLTWIVGAAG VMESFLIVAM C CTCTMLTA ISMSAIATNG VVPAGGSYYM ISRSLGPEFG GAVGLCFYLG TTFAGAMYIL GTIEIFLTYI SPSAAIFQAE TA DGEAAAL LNNMRVYGSC ALALMAVVVF VGVKYVNKLA LVFLACVVLS ILAIYAGVIK TAFAPPDIPV CLLGNRTLAN RNF DTCAKM QVVSNGTVTT ALWRLFCNGS SLGATCDEYF AQNNVTEIQG IPGVASGVFL DNLWSTYSDK GAFVEKKGVS SVPV SEESR PGGLPYVLTD IMTYFTMLVG IYFPSVTGIM AGSNRSGDLK DAQKSIPTGT ILAIVTTSFI YLSCIVLFGA CIEGV VLRD KFGEALQGNL VIGMLAWPSP WVIVIGSFFS TCGAGLQSLT GAPRLLQAIA RDGIIPFLQV FGHGKANGEP TWALLL TAL ICETGILIAS LDSVAPILSM FFLMCYMFVN LACAVQTLLR TPNWRPRFKF YHWTLSFLGM SLCLALMFIC SWYYALF AM LIAGCIYKYI EYRGAEKEWG DGIRGLSLNA ARYALLRVEH GPPHTKNWRP QVLVMLNLDS EQCVKHPRLL SFTSQLKA G KGLTIVGSVL EGTYLDKHVE AQRAEENIRS LMSAEKTKGF CQLVVSSNLR DGASHLIQSA GLGGMKHNTV LMAWPEAWK EADNPFSWKN FVDTVRDTTA AHQALLVAKN IDLFPQNQER FSDGNIDVWW IVHDGGMLML LPFLLRQHKV WRKCRMRIFT VAQVDDNSI QMKKDLQMFL YHLRISAEVE VVEMVENDIS AFTYEKTLMM EQRSQMLKQM QLSKNERERE AQLIHDRNTA S HTTATART QAPPTPDKVQ MTWTKEKLIA EKHRNKDTGP SGFKDLFSLK PDQSNVRRMH TAVKLNGVVL NKSQDAQLVL LN MPGPPKS RQGDENYMEF LEVLTEGLNR VLLVRGGGRE VITIYSSNSL EVLFQ UniProtKB: Solute carrier family 12 member 7 |
-Macromolecule #3: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #4: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3uL, 1 blot force, 4s blot time, 1s wait time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 0.11 sec. / Average electron dose: 0.9333 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 36000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















